Welcome to INSaFLU’s documentation!

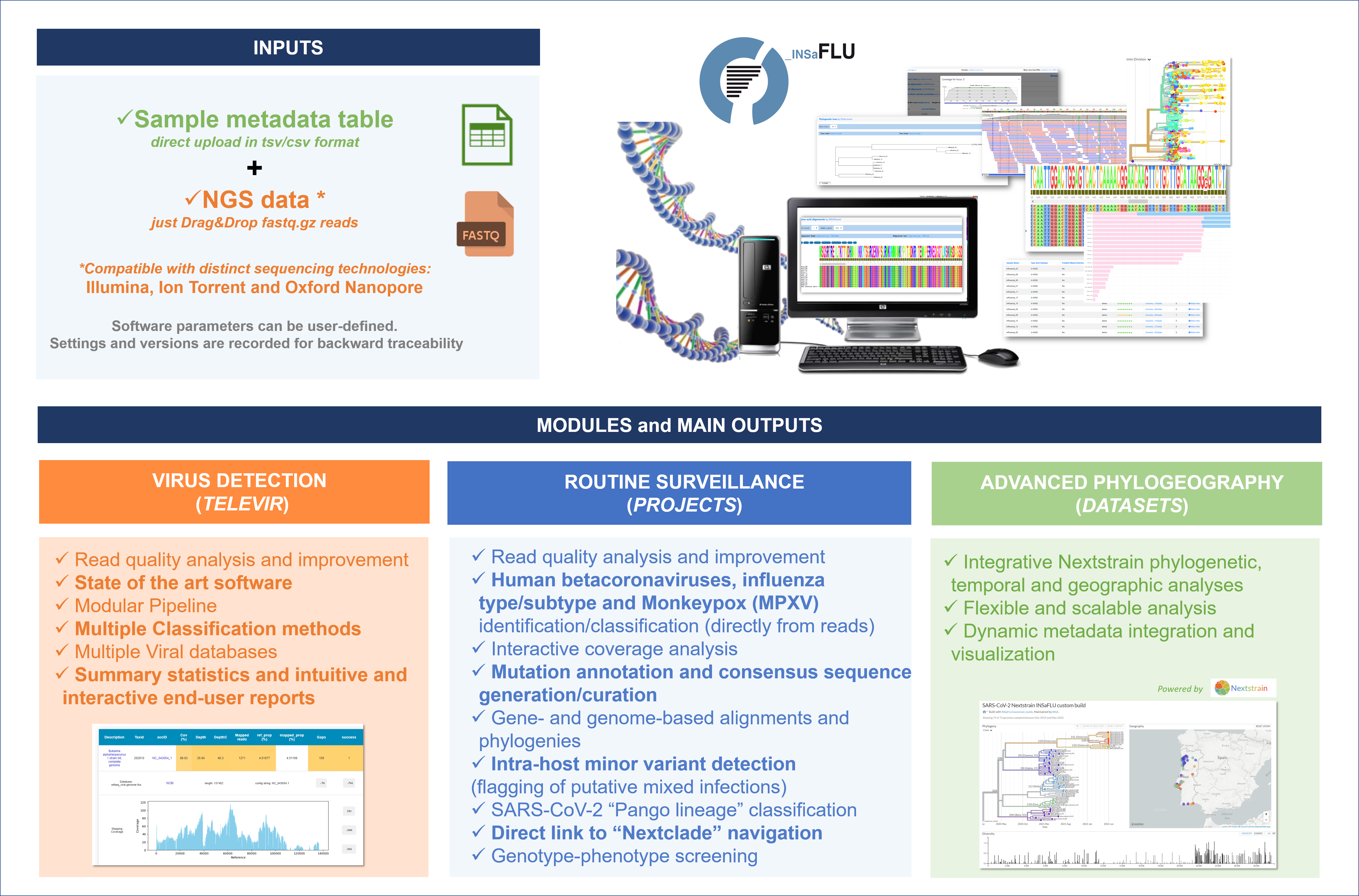
INSaFLU-TELEVIR platform is a free bioinformatics web-based (but also locally installable) suite that deals with primary sequencing data (Illumina, Ion Torrent and Oxford Nanopore Technologies reads) towards:
metagenomics virus detection (from reads to virus detection)
routine genomic surveillance (from reads to mutations detection, consensus generation, virus classification, alignments, “genotype-phenotype” screening, phylogenetics, integrative Nextstrain phylogeographical and temporal analysis etc).
INSaFLU-TELEVIR versatility and functionality is expected to supply public health laboratories and researchers with a user-oriented “start-to-end” bioinformatics framework that can potentiate a strengthened and timely detection and monitoring of viral (emerging) threats.
Online tool: https://insaflu.insa.pt
Documentation / Tutorial: https://insaflu.readthedocs.io/en/latest/
Easy local installation: https://github.com/INSaFLU/docker
Overview
How to Cite
If you use INSaFLU in your work, please cite these publications:
Borges V, Pinheiro M et al. Genome Medicine (2018) 10:46 https://doi.org/10.1186/s13073-018-0555-0
Dourado Santos et al, pre-prrint (Research Square) https://doi.org/10.21203/rs.3.rs-3556988/v1
Contact
If you have any questions, comments or suggestions, please contact us: <vitor.borges@insa.min-saude.pt> or <insaflu@insa.min-saude.pt>