Reference-based genomic surveillance
The surveillance-oriented component of INSaFLU allows running:
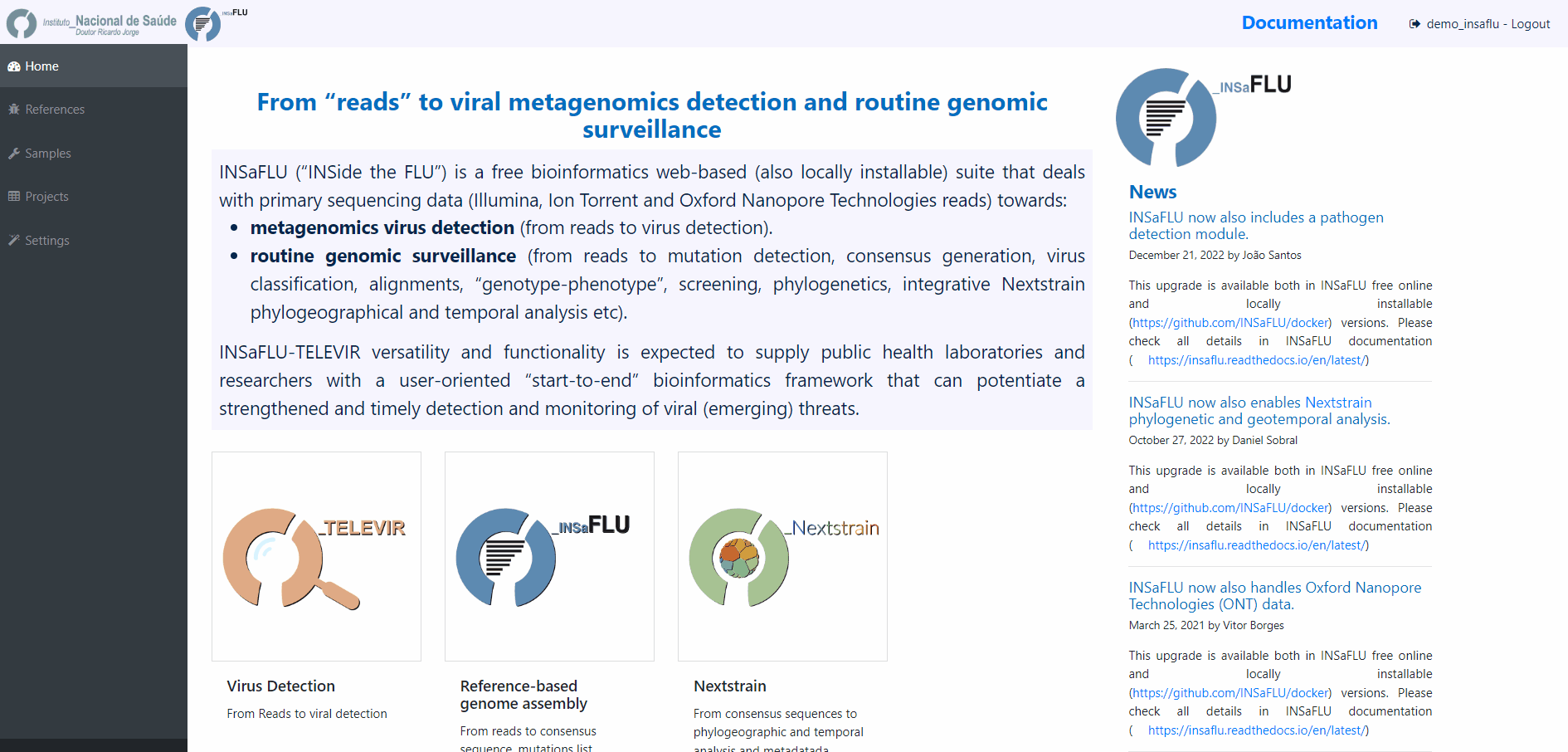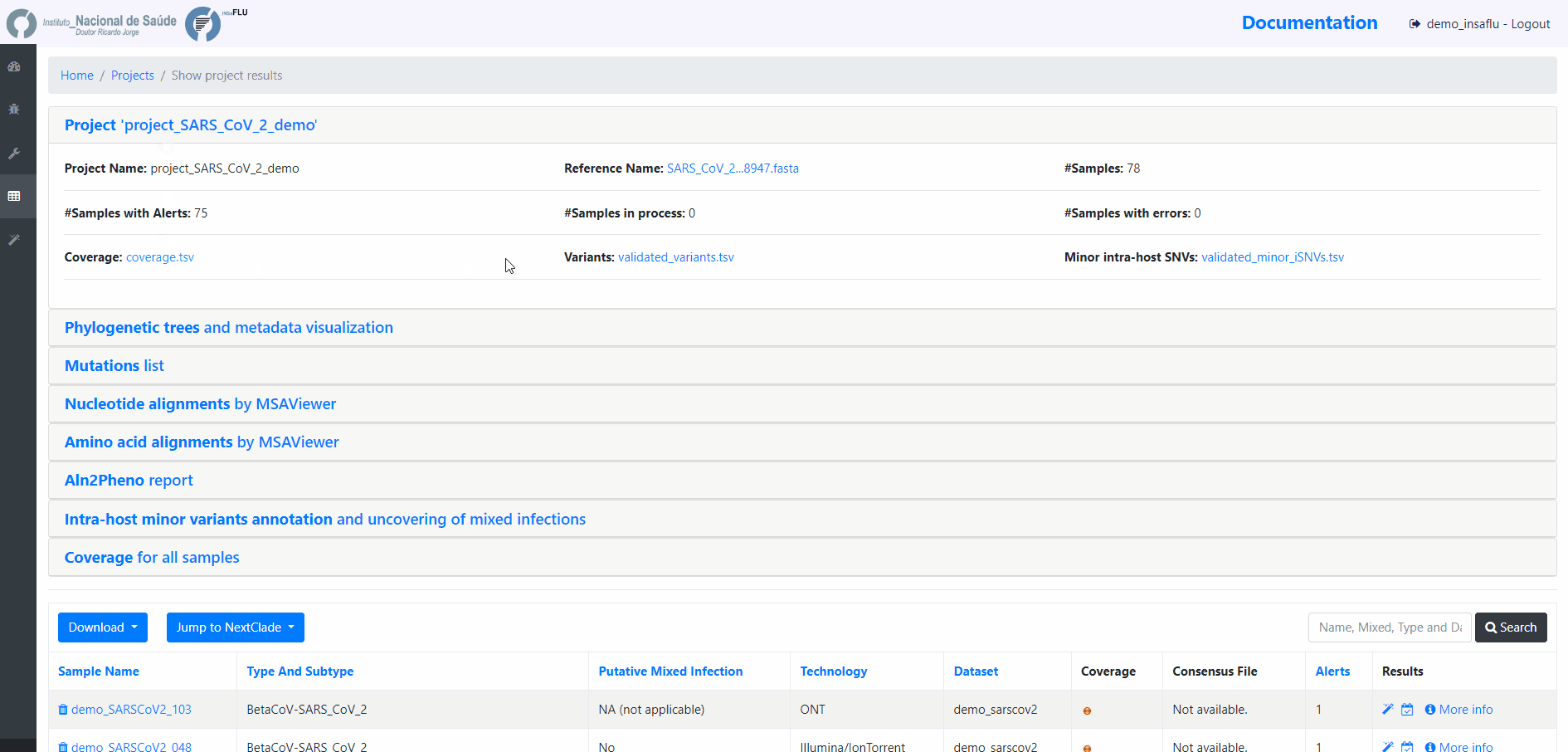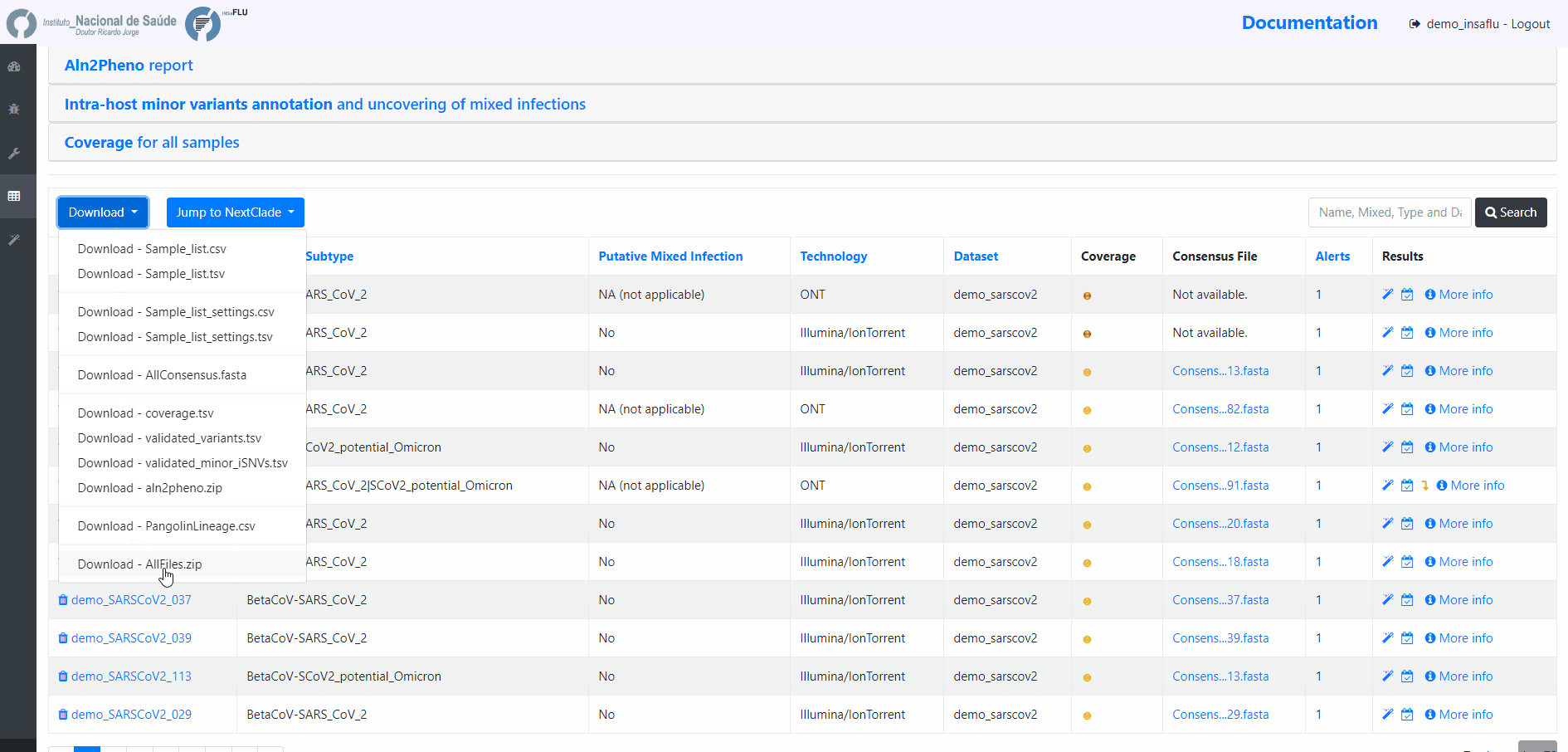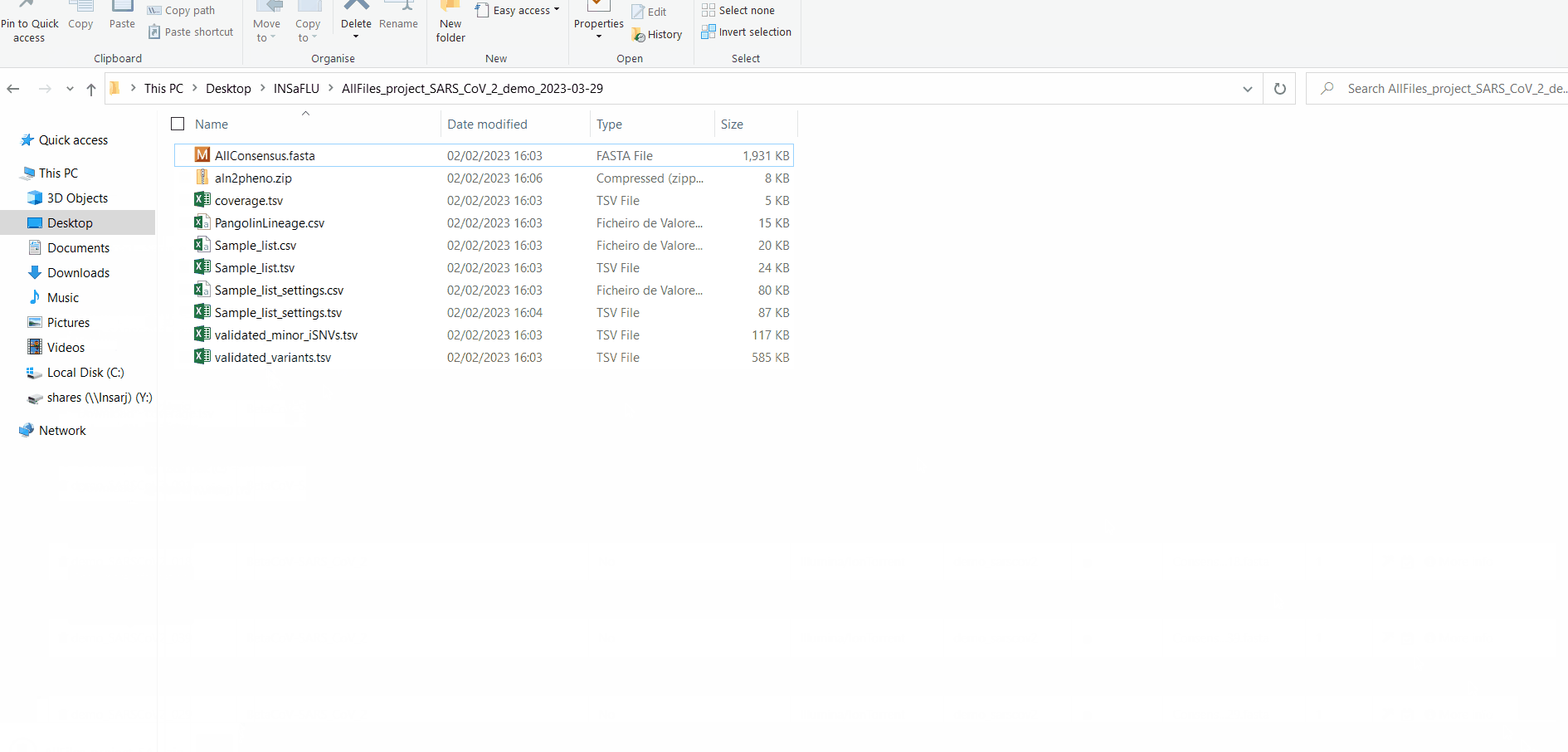
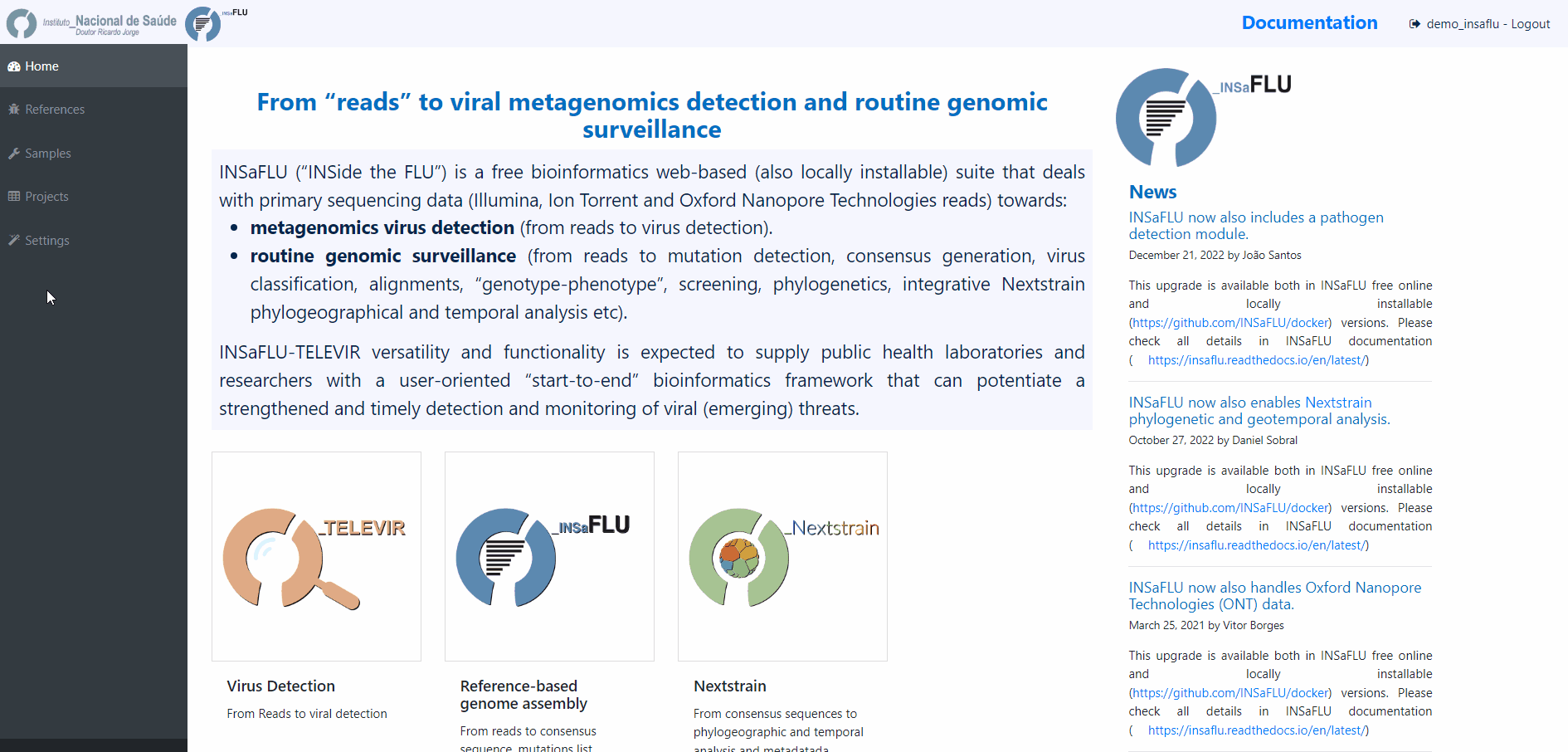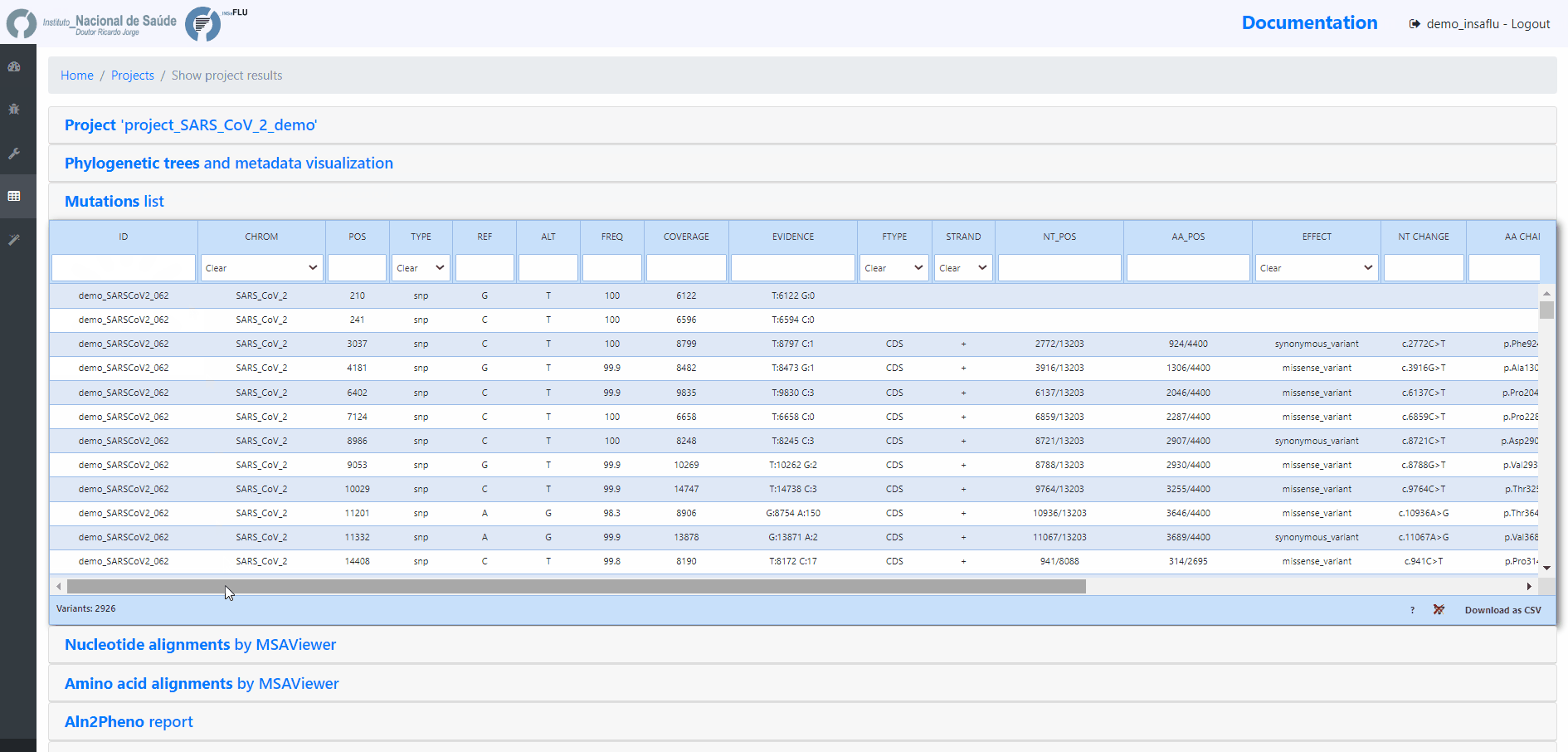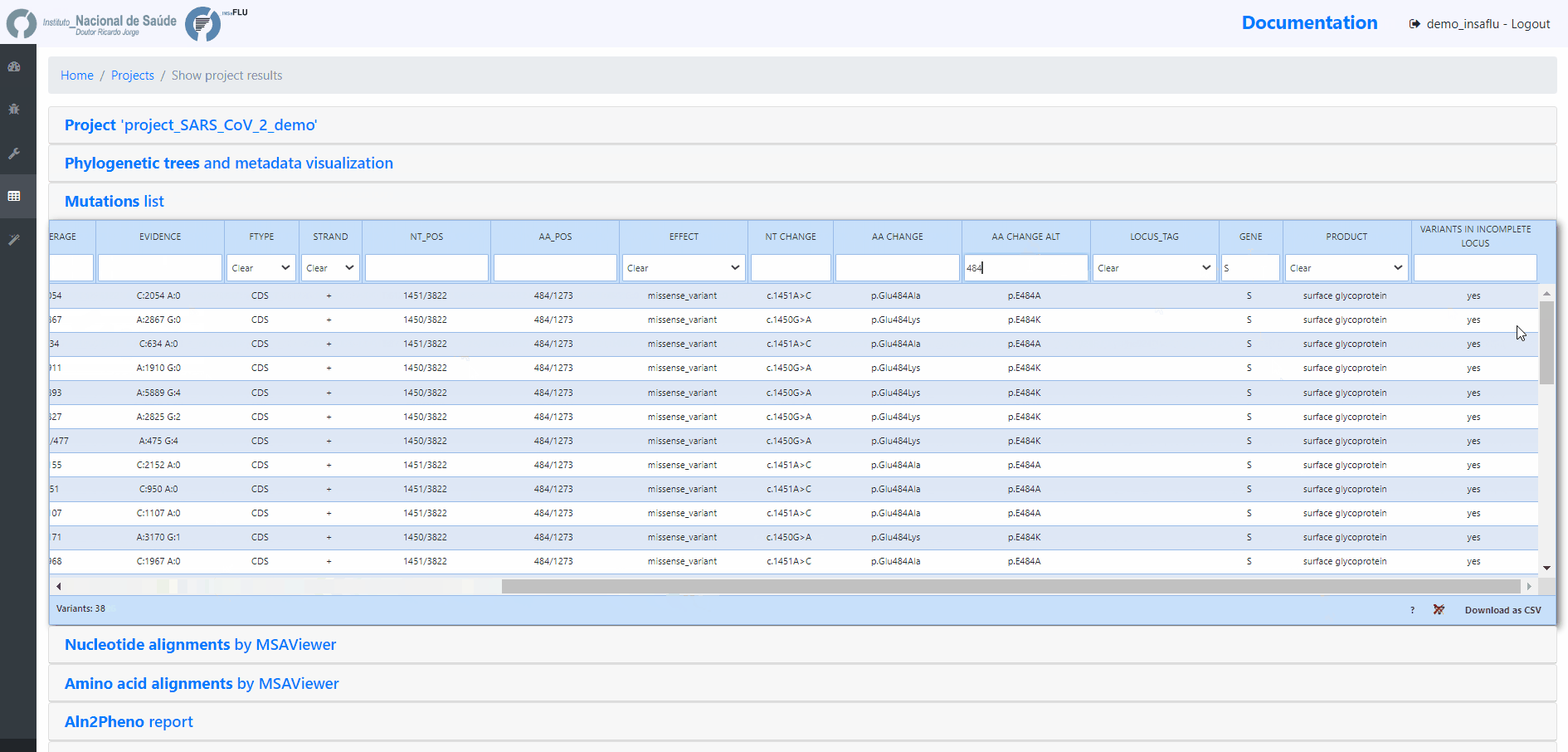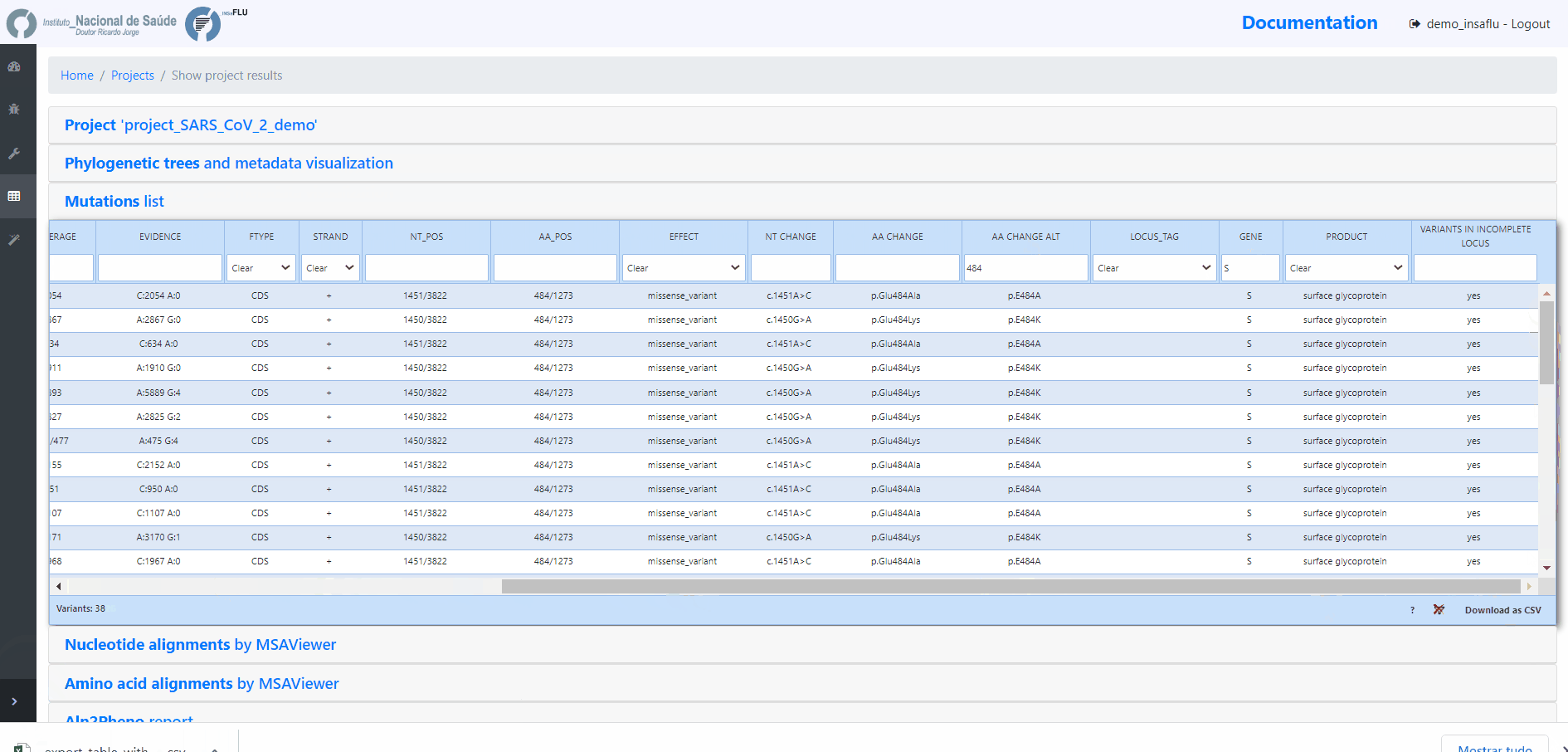
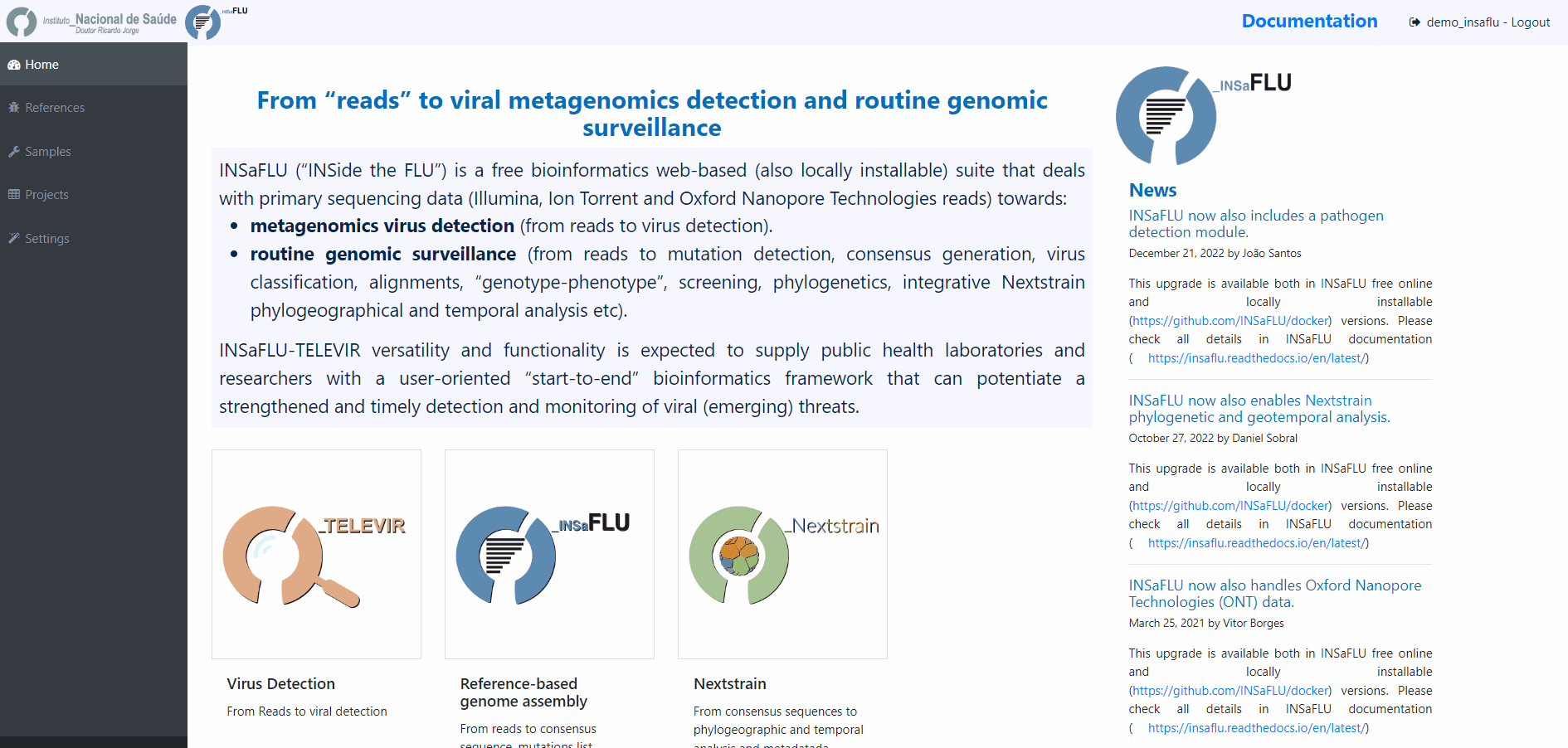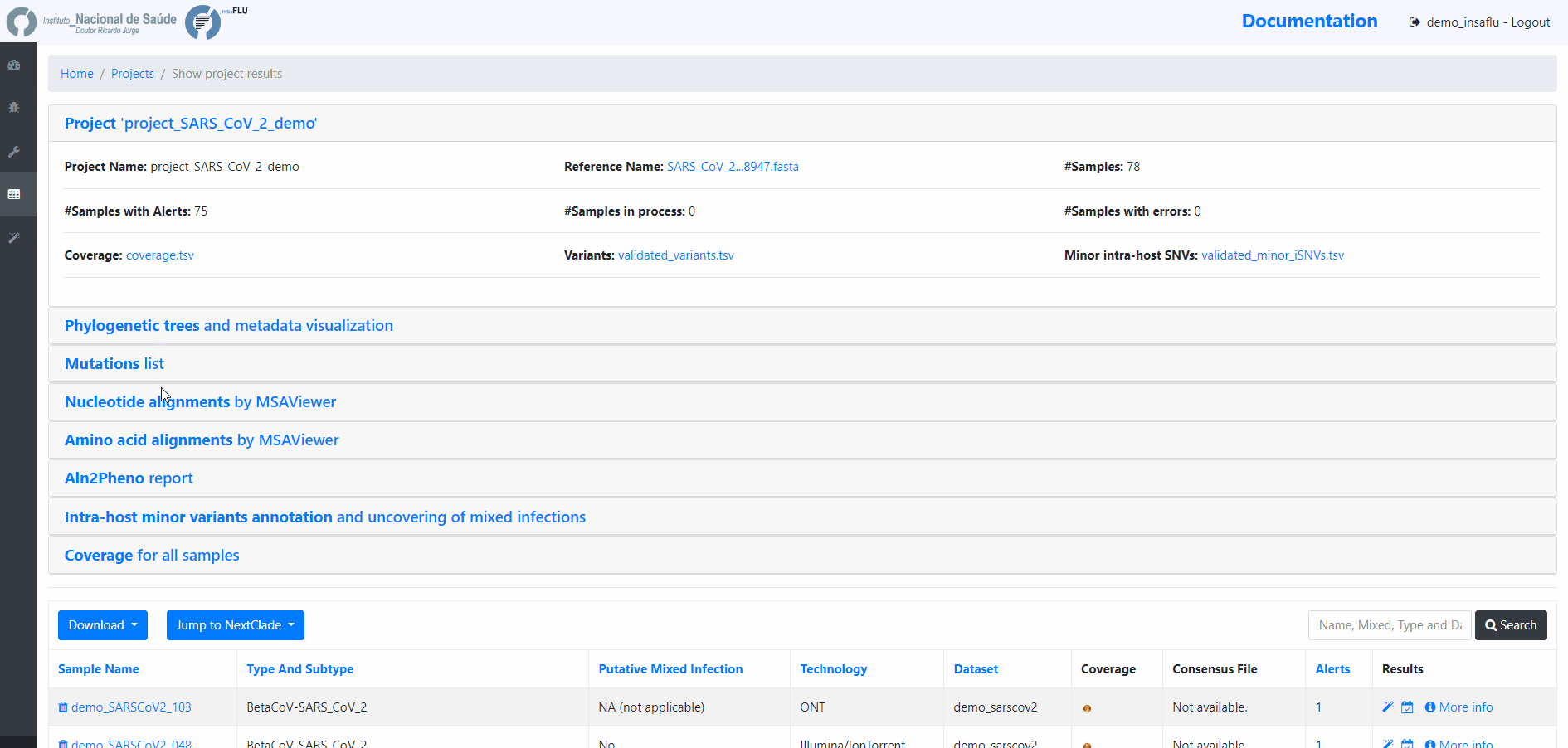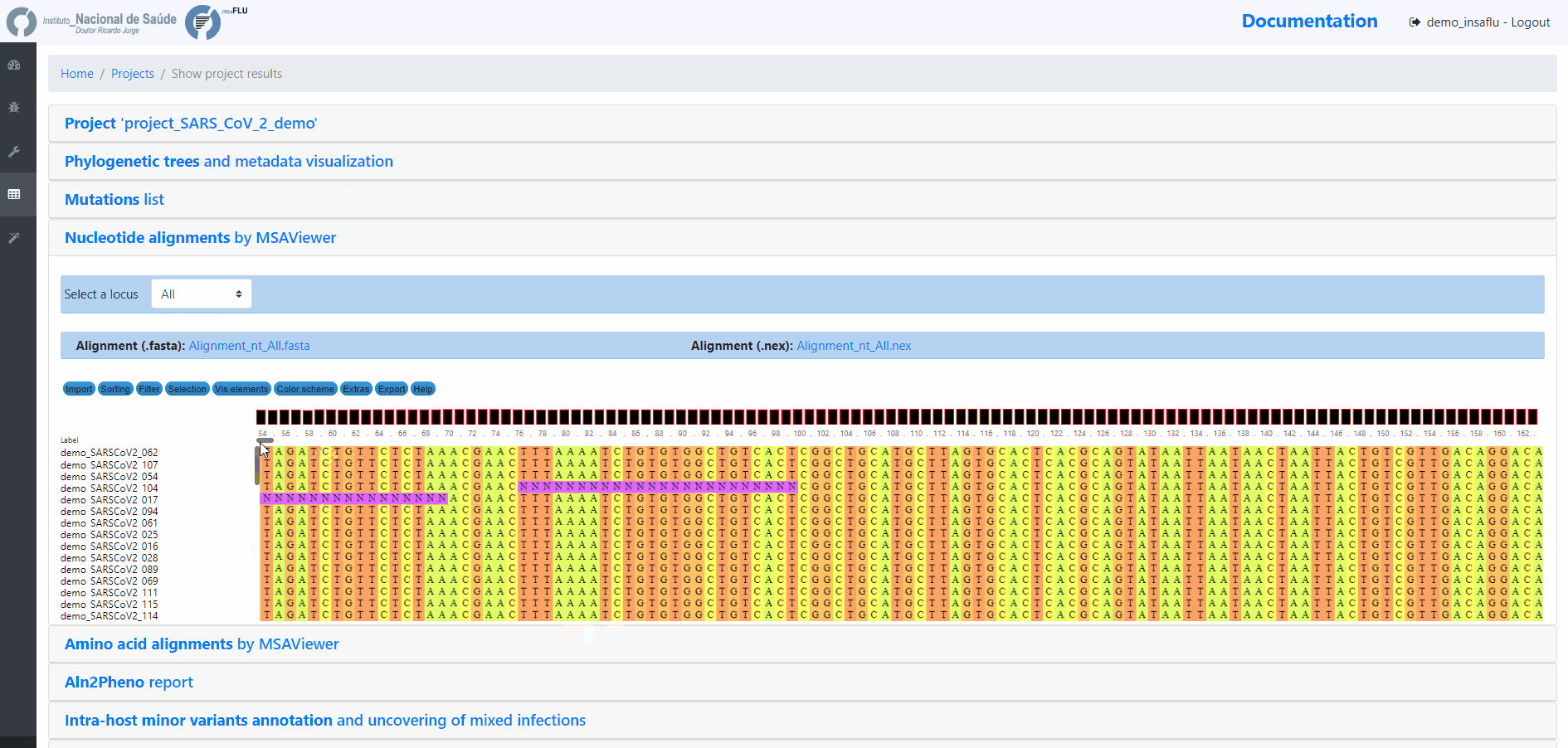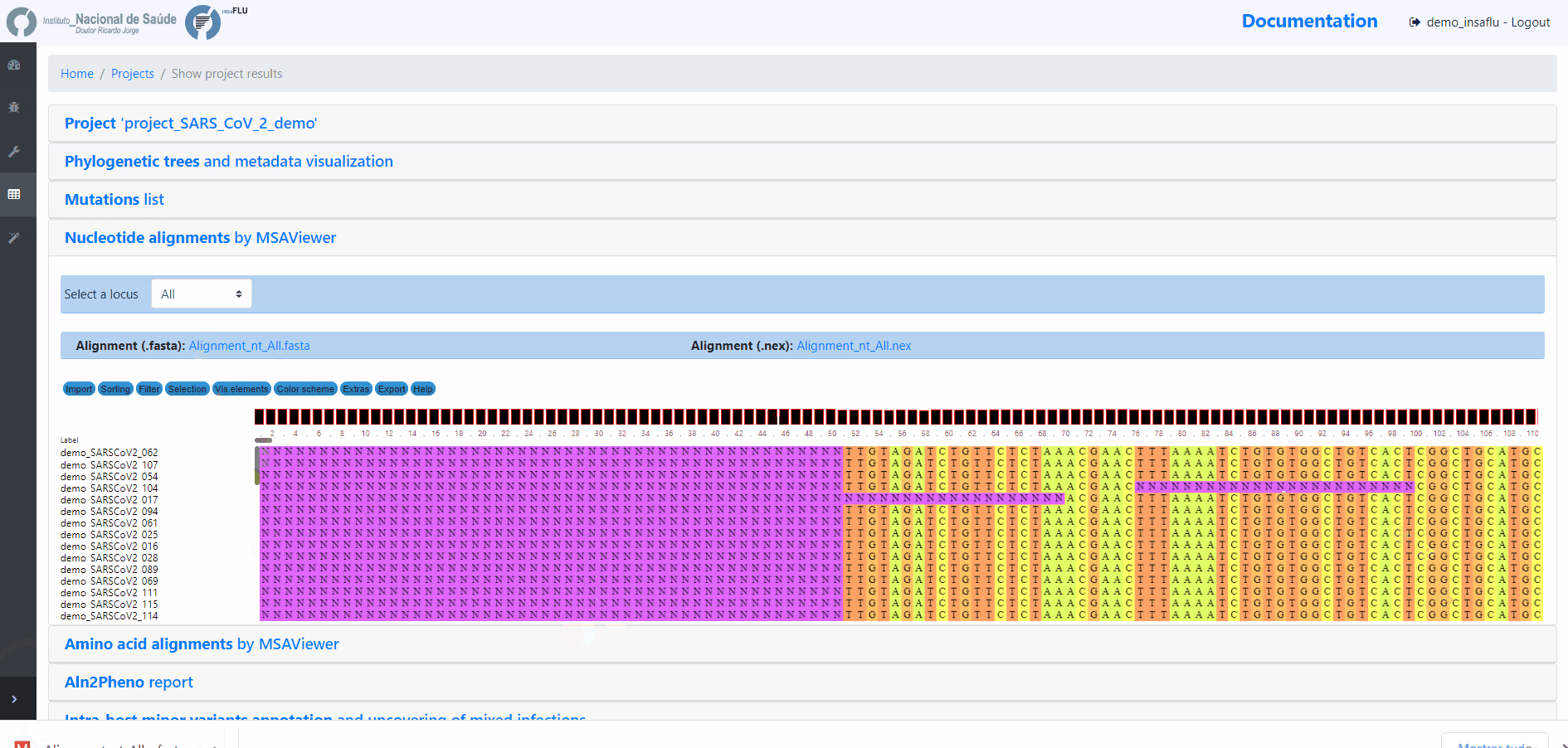
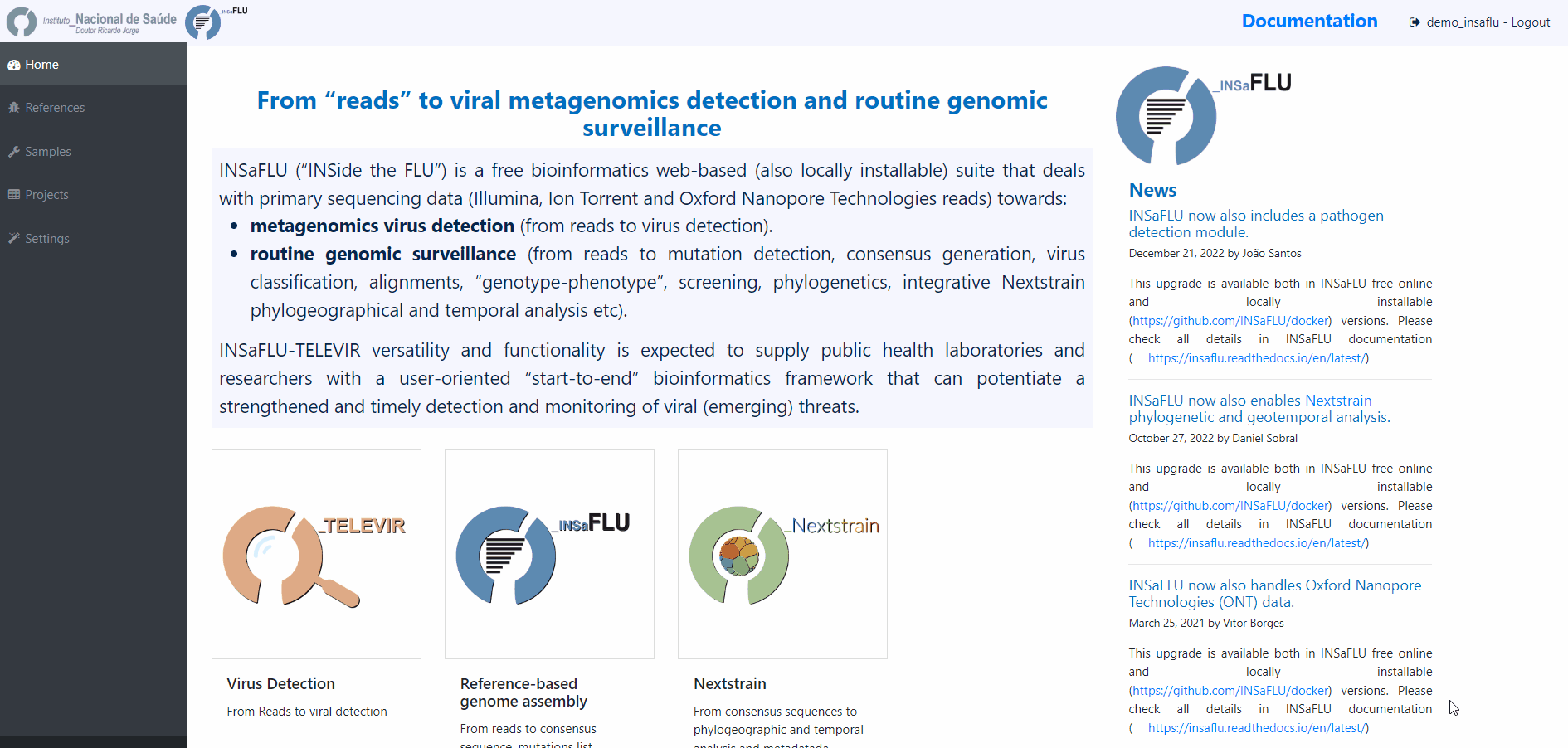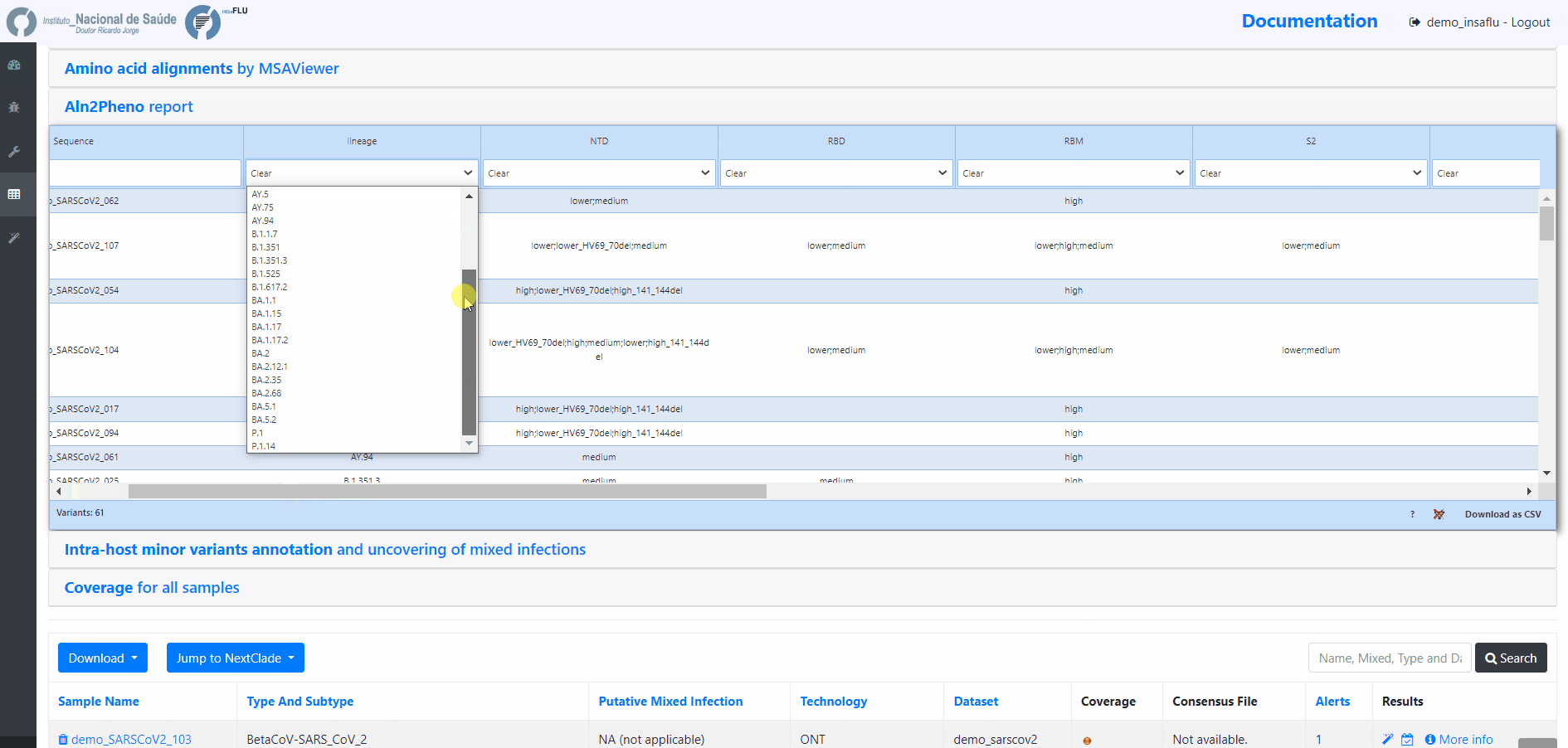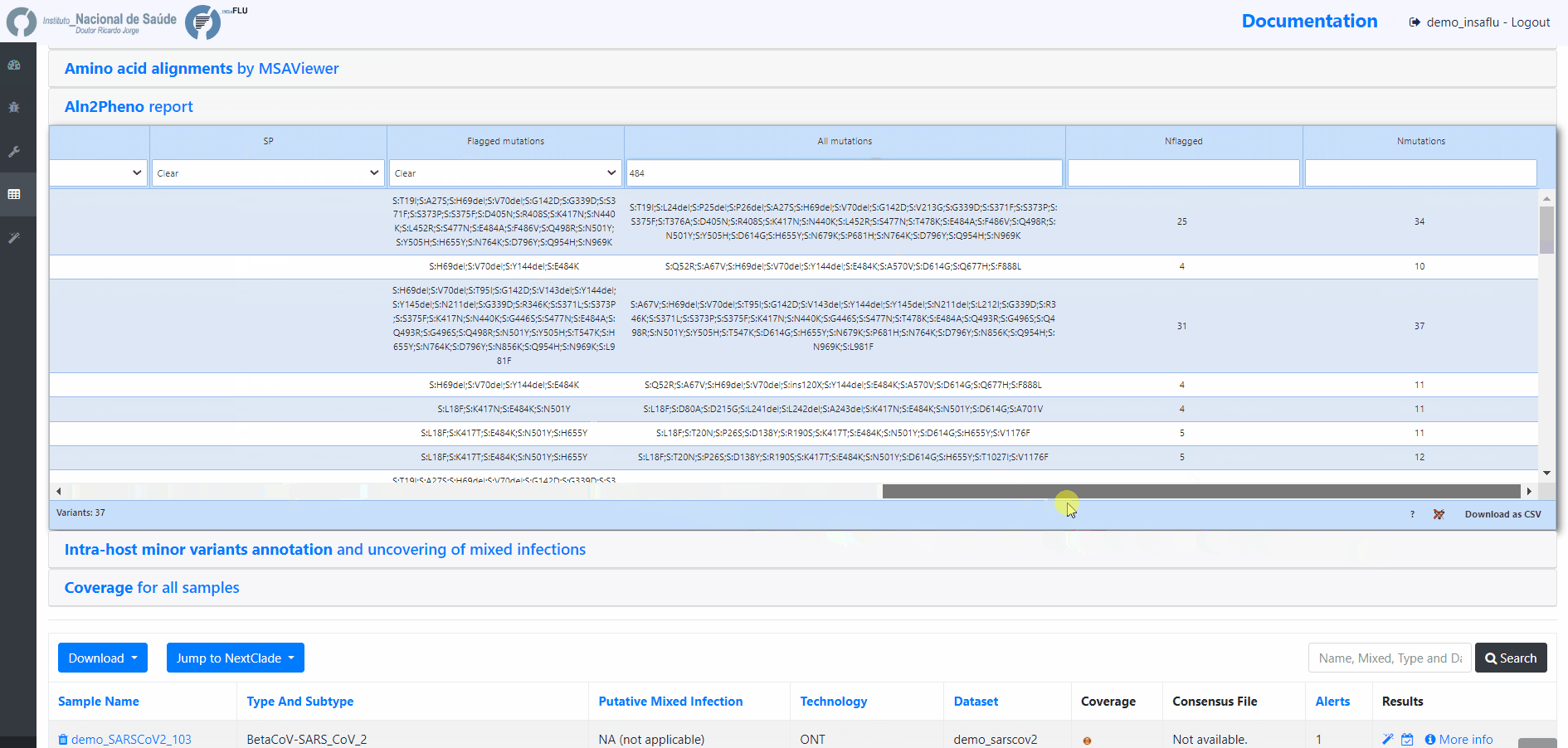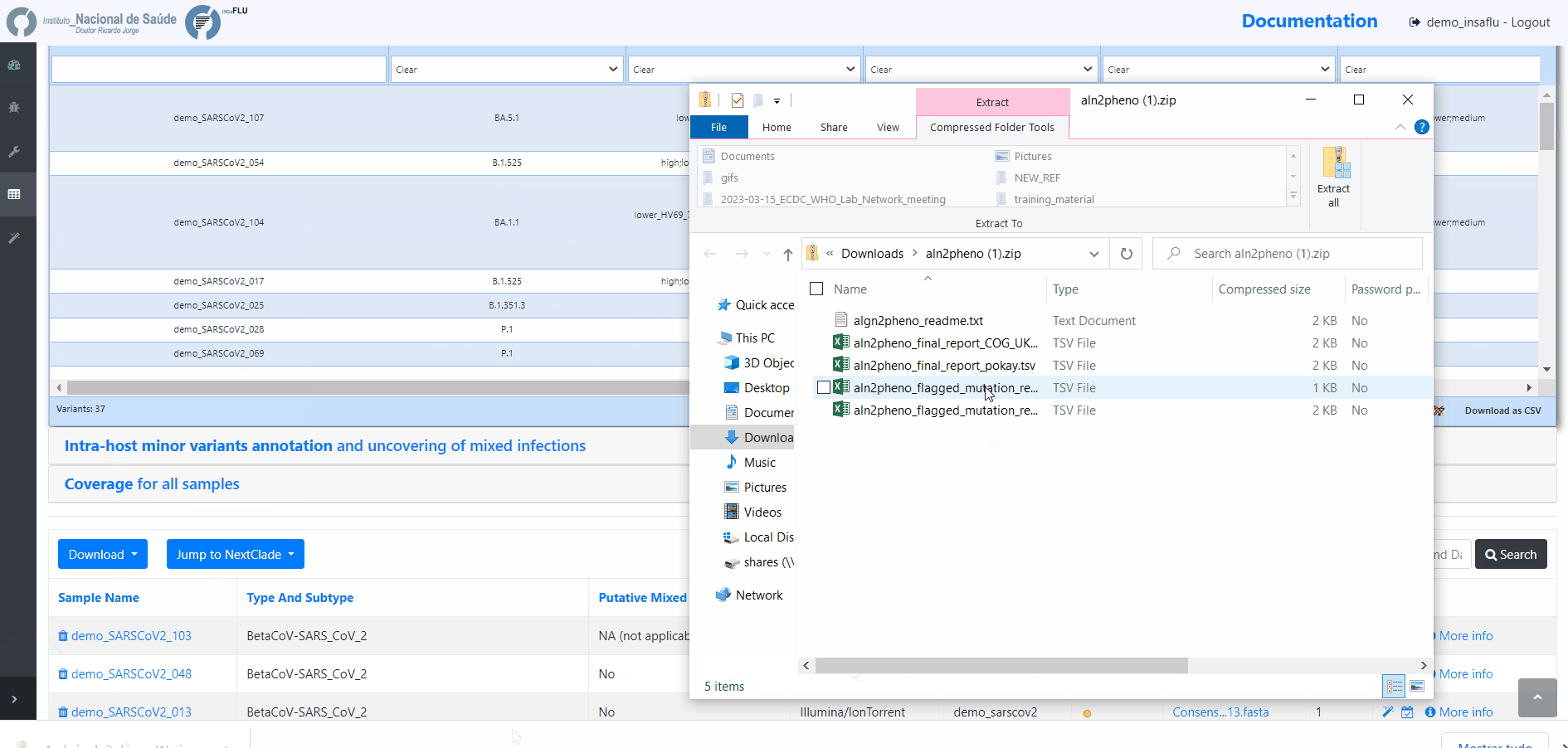
INSaFLU Projects - From reads to reference-based generation of consensus sequences and mutations annotation, followed by gene- and genome-based alignments, amino acid alignments, Pango classification, NextClade linkage, etc.
Nextstrain Datasets - From consensus sequences to advanced Nextstrain phylogenetic and genomic analysis, coupled with geographic and temporal data visualization and exploration of sequence metadata.
One of the main goals of INSaFLU is to make data integration completely flexible and scalable in order to fulfill the analytical demands underlying routine genomic surveillance throughout viral epidemics. As such, INSaFLU allows users to create several projects or datasets and add more samples to each one as needed. In a dynamic manner, project / dataset outputs are automatically re-build and cumulatively updated as more samples are added to each project / dataset. The outputs are provided to be compatible with multiple downstream applications.
INSaFLU Projects - How to create and scale-up a genomic surveillance project
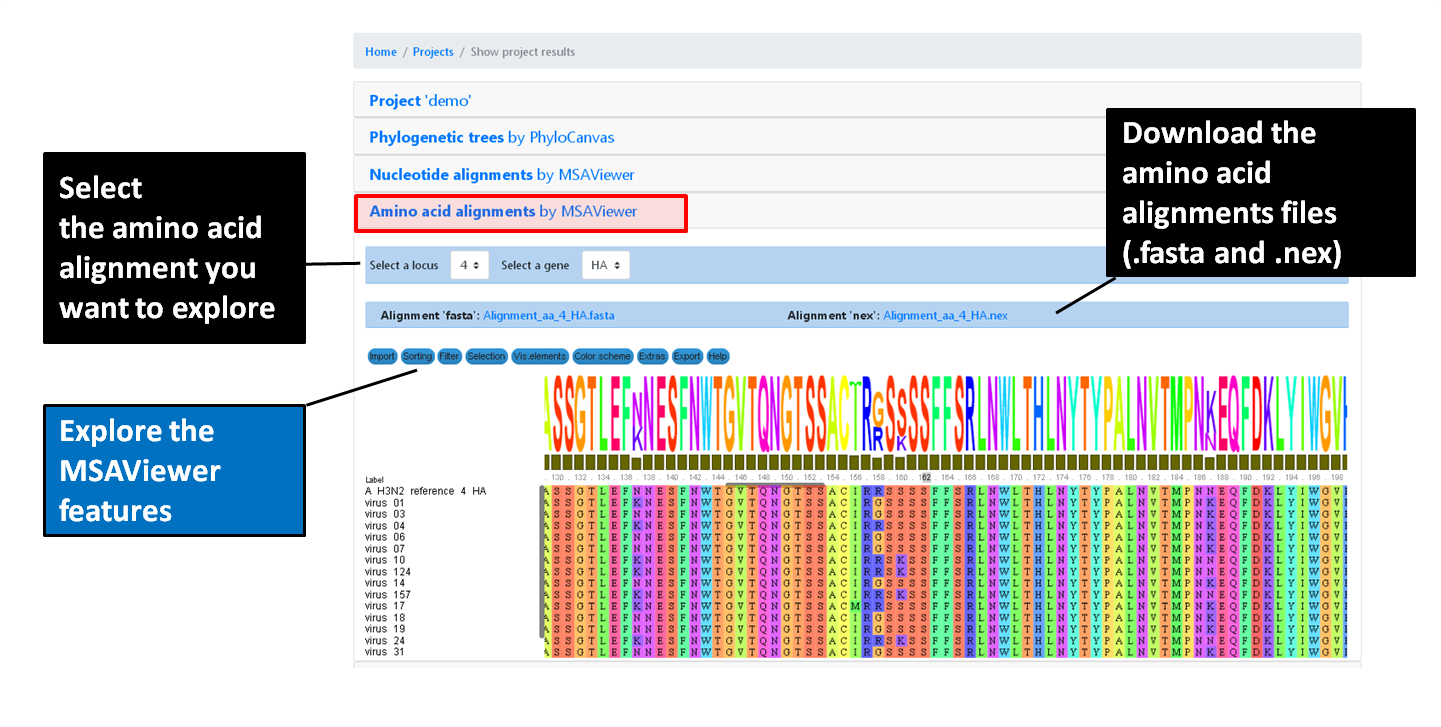
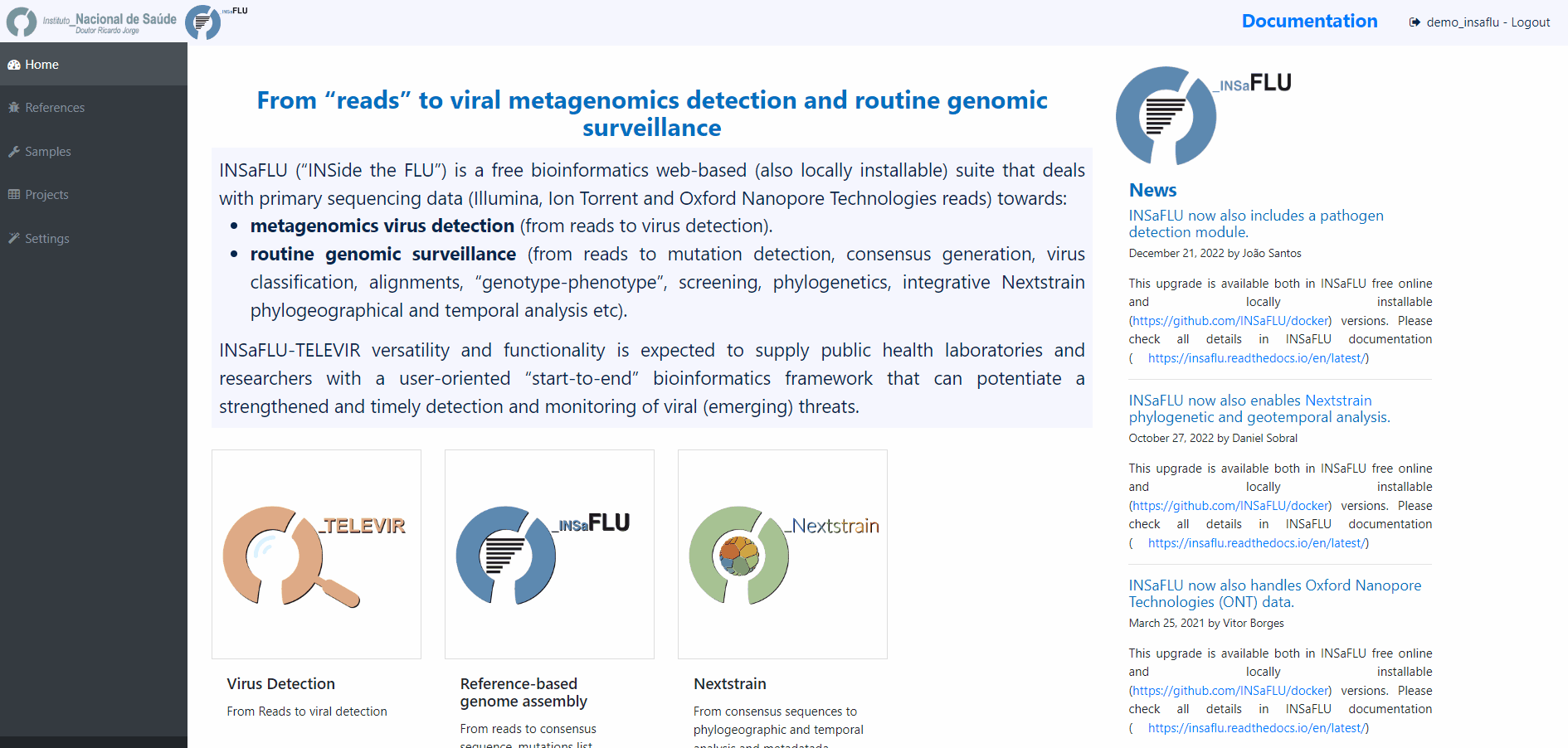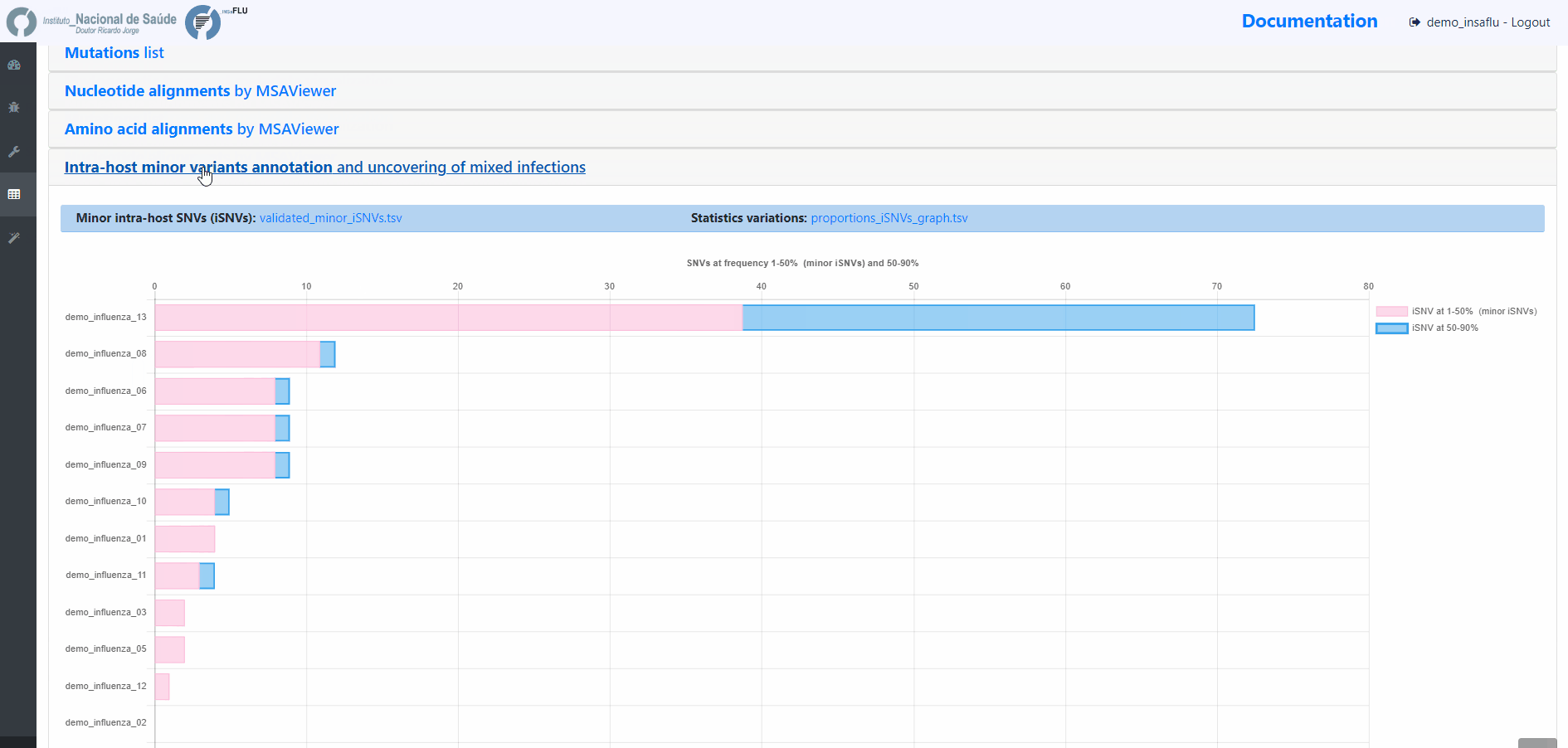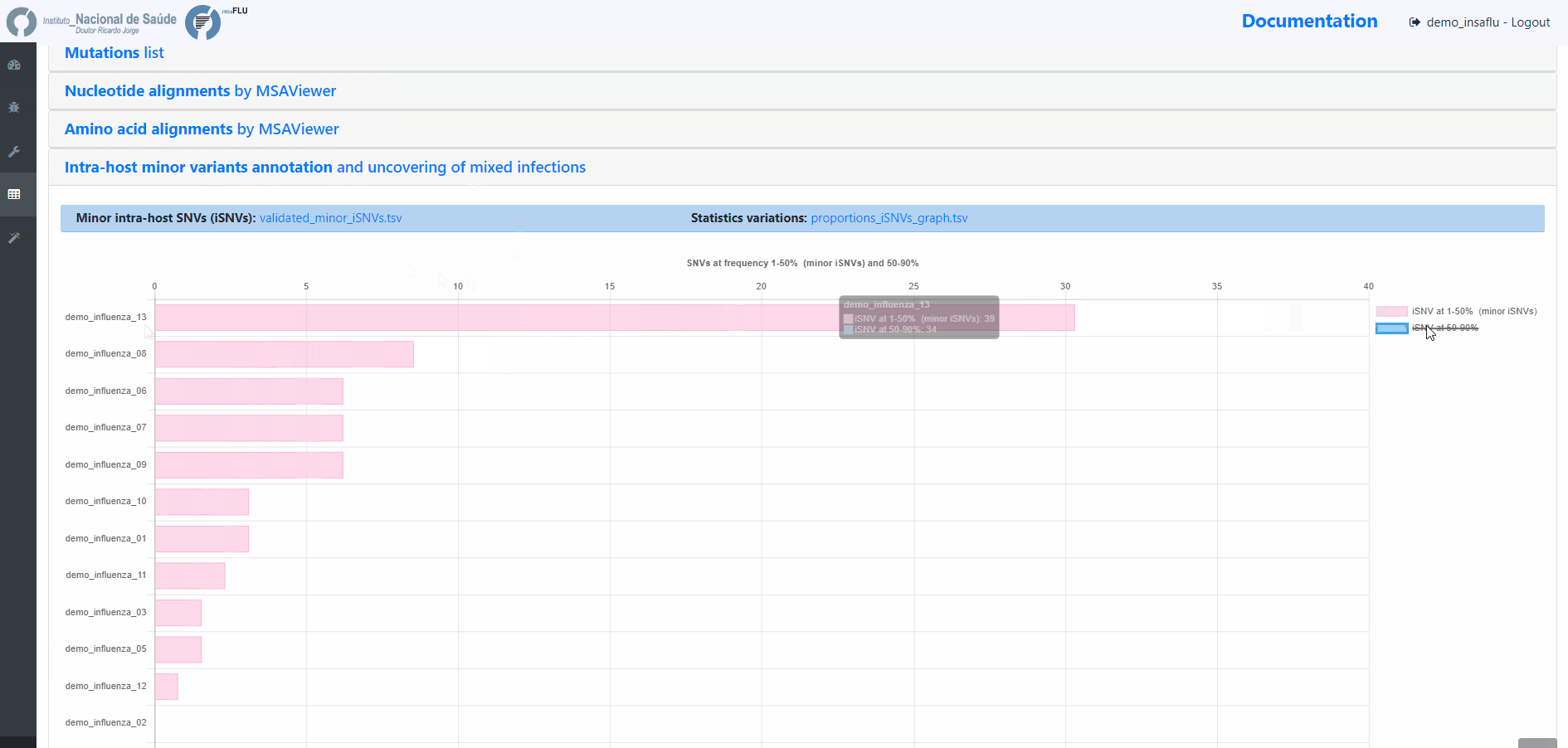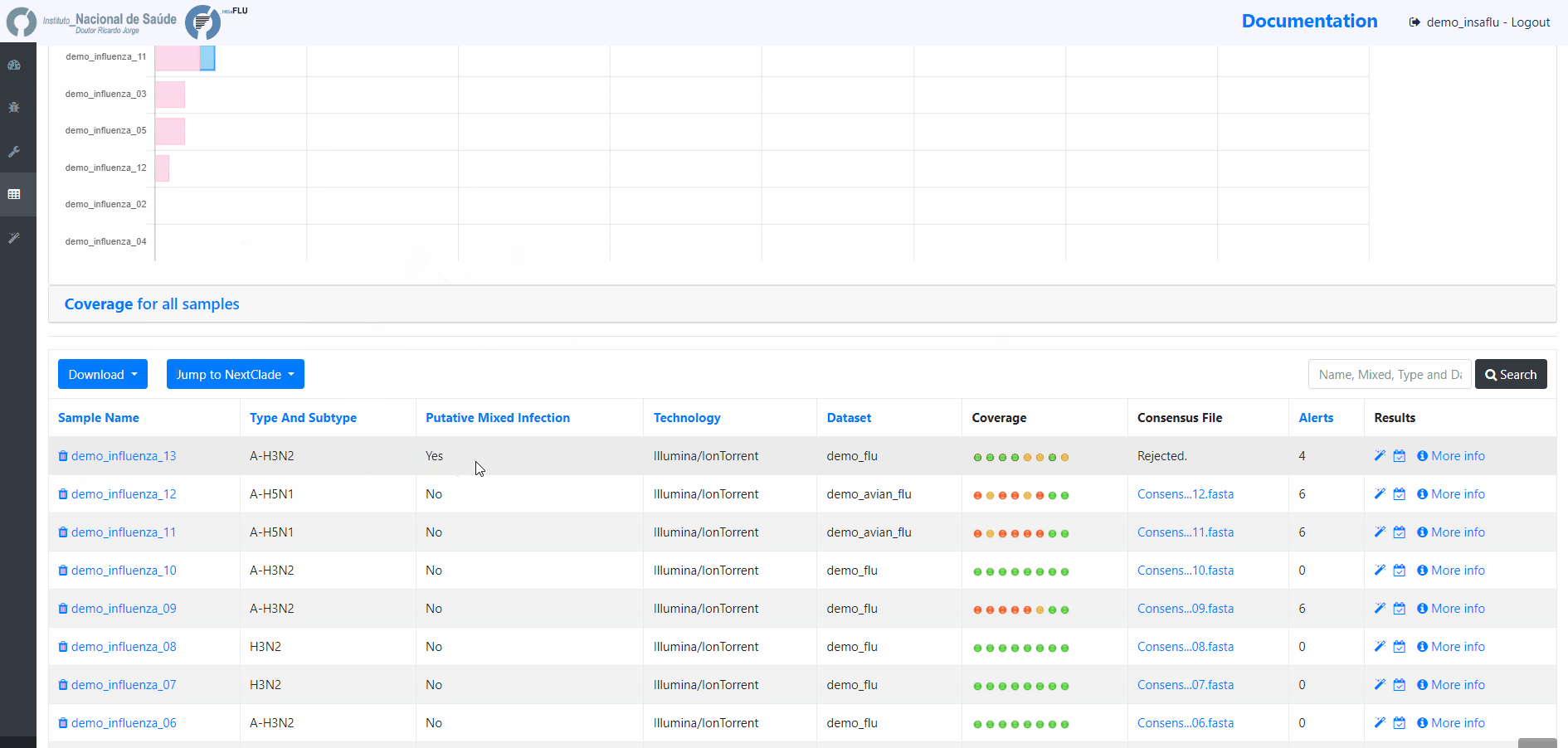
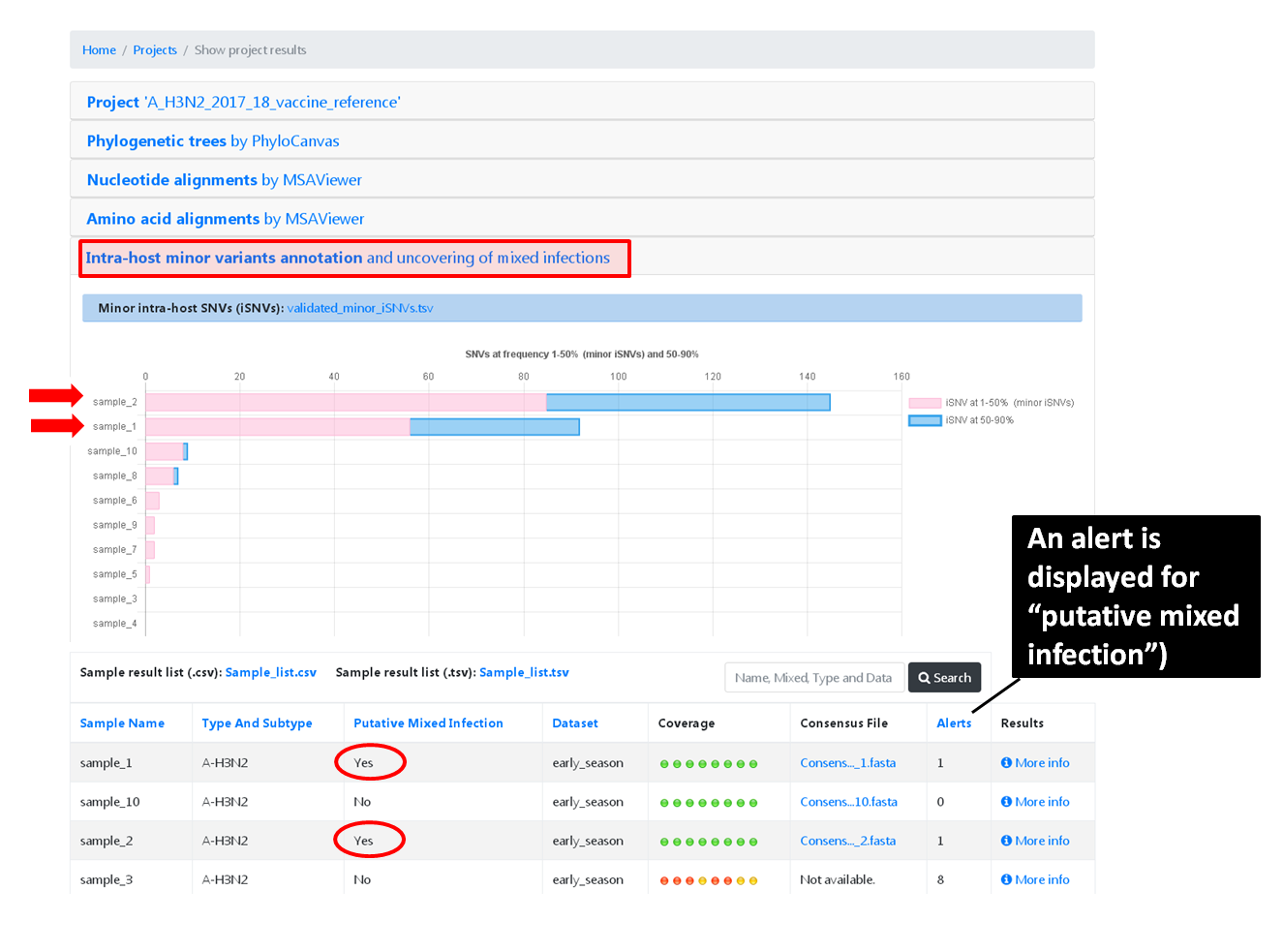
Projects - From reads to reference-based generation of consensus sequences and mutations annotation, followed by gene- and genome-based alignments, amino acid alignments, Pango classification, NextClade linkage, etc.
Within the Projects menu:
2. Choose a Project Name, select a Reference sequence and Save
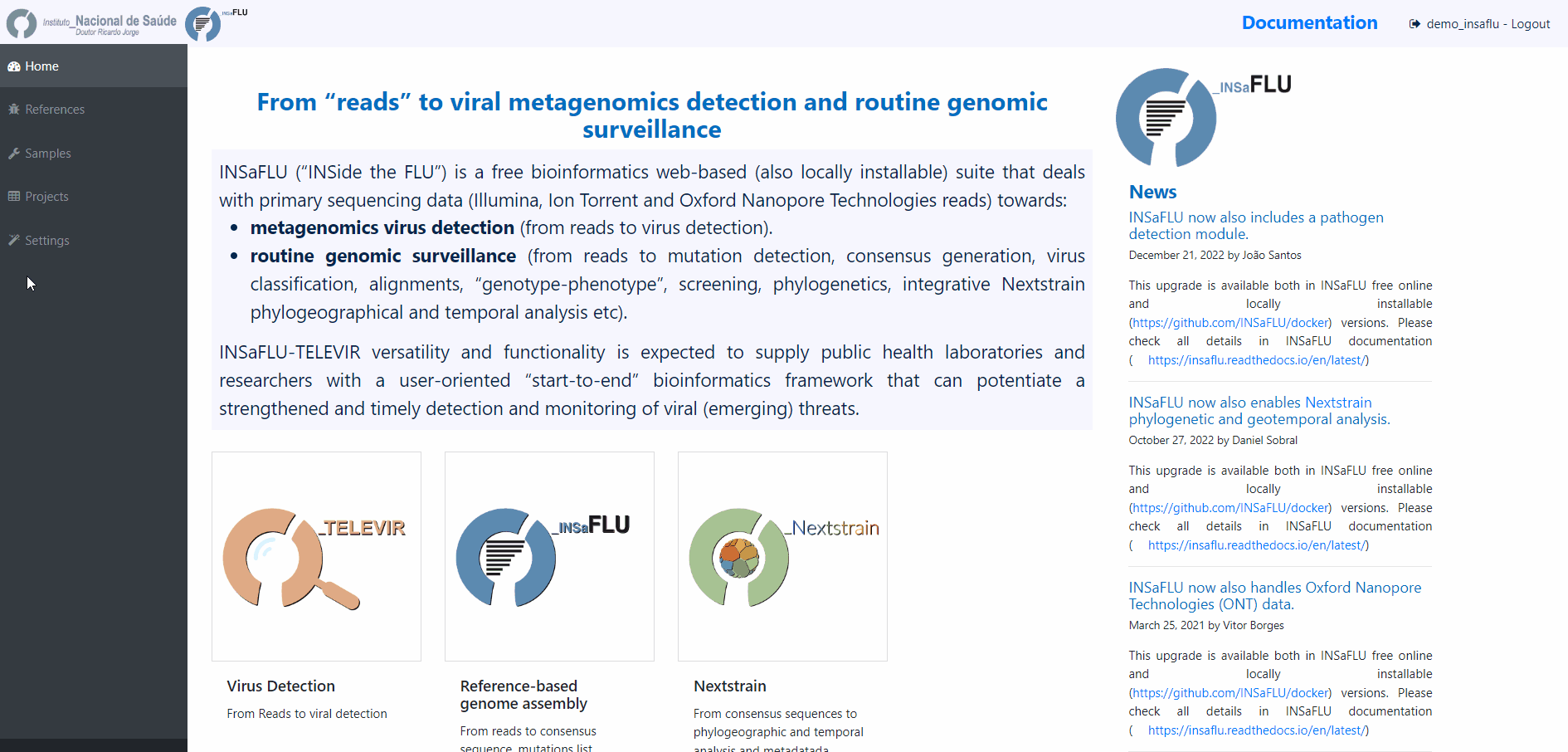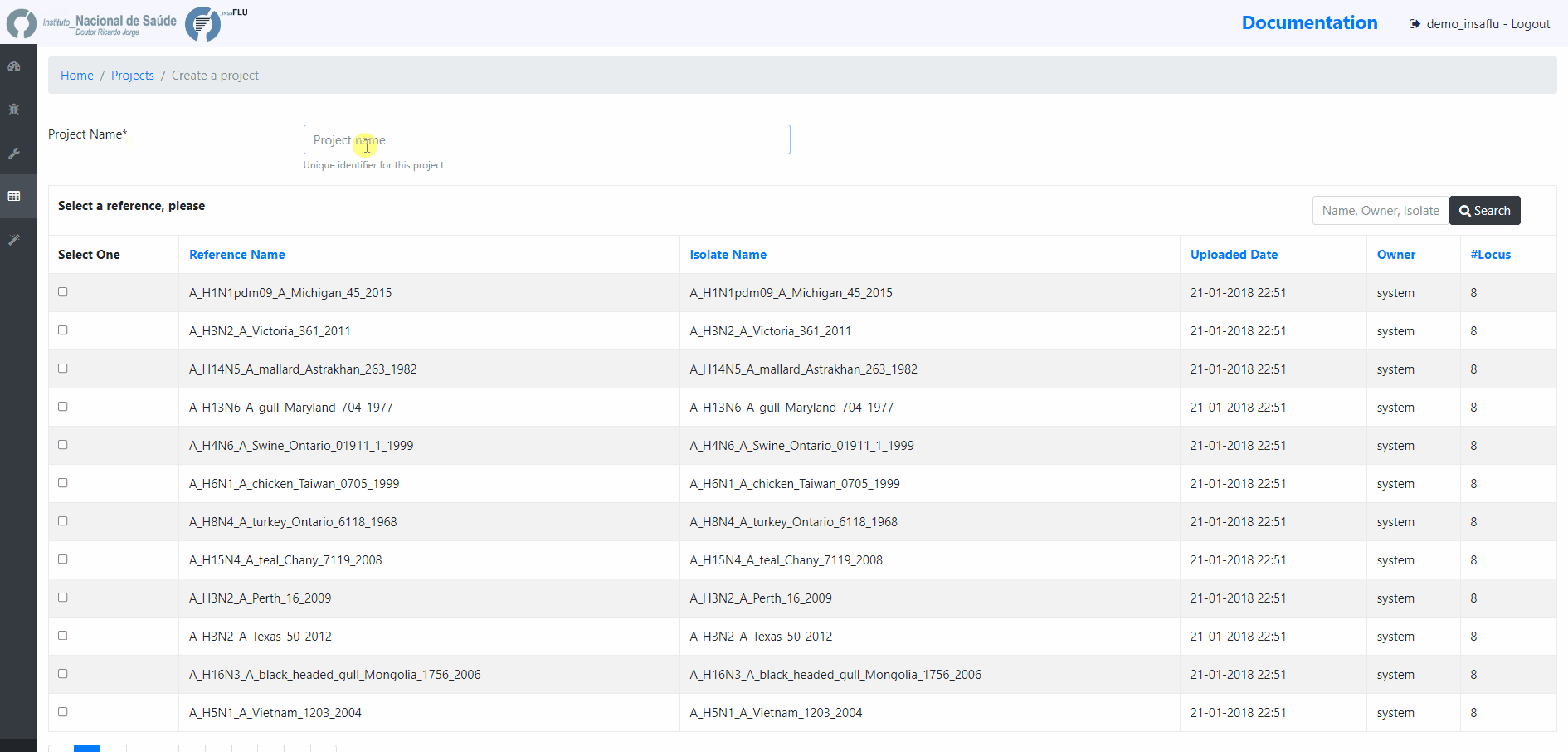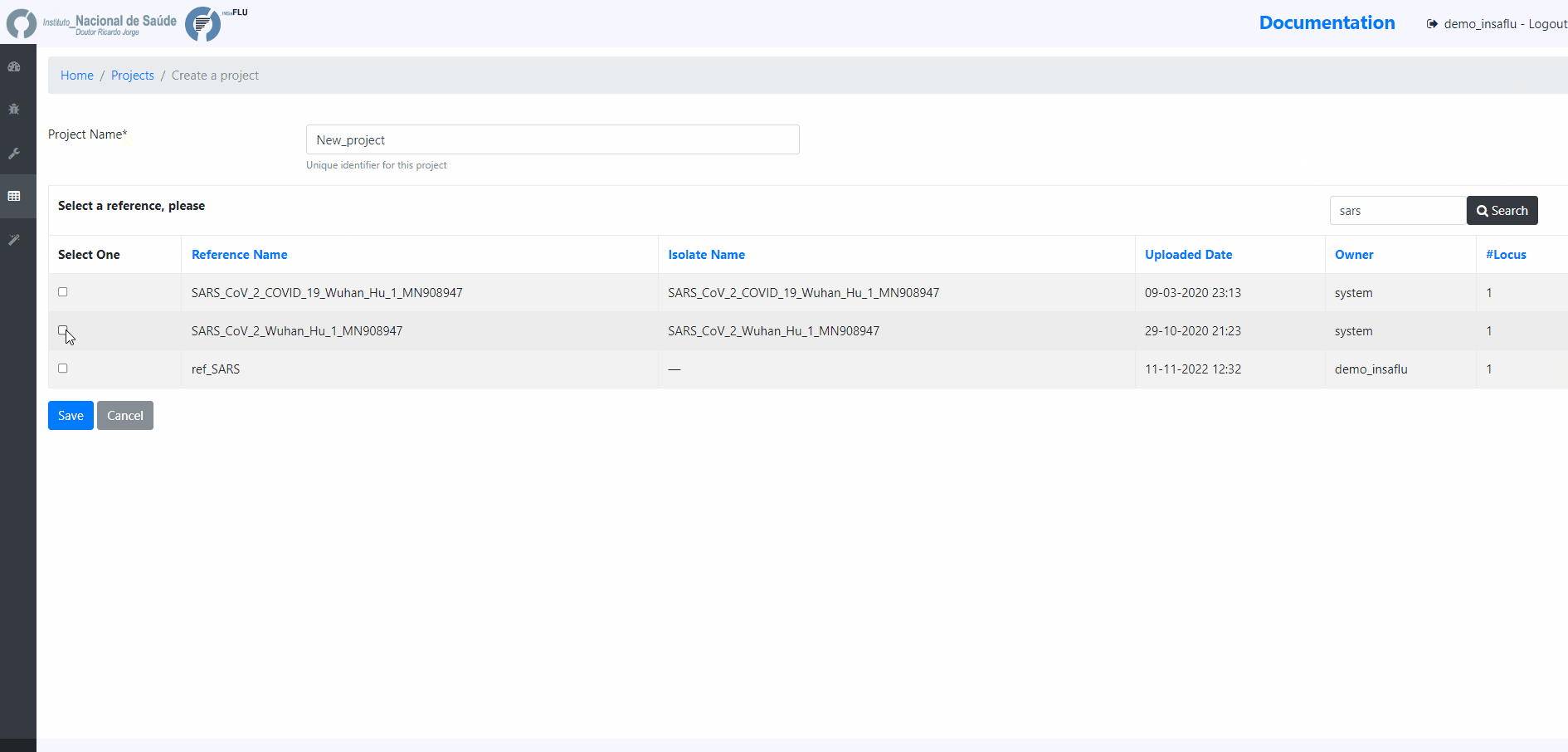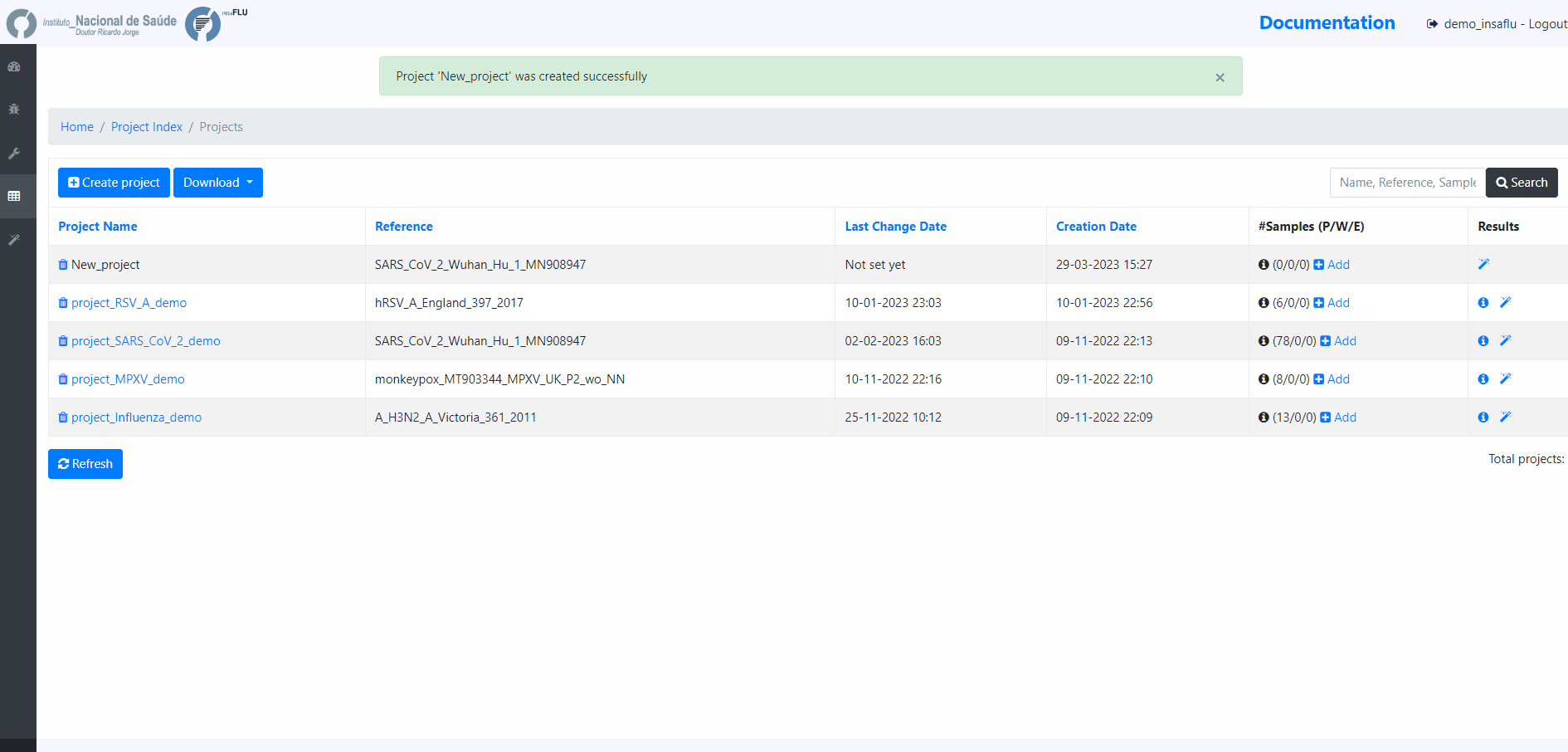
You are encouraged to create “umbrella” projects, such as projects enrolling same sub-type viruses from the same season that will be compared with the vaccine reference virus for a given flu season.
You can designate the projects so that the name easily indicates the combination “virus sub-type/season/reference” (e.g. A_H3N2_2017_18_vaccine_ref)
Important
You should select a reference sequence (e.g., the vaccine strain from the current influenza season) that fits both your amplicon design (i.e., a multi-fasta file containing the set of reference sequences with the precise size of each “intra-amplicon” target sequence that you capture by each one of the RT-PCR amplicons) and the set of samples that will be compared (e.g., same sub-type viruses from the same season to be compared with the vaccine reference virus).
3. Choose the software parameters to be applied to the project.
After creating a project, and before adding the first sample, you can clicking in the “Magic wand” to select the parameters to be applied by default to every sample added to the project.
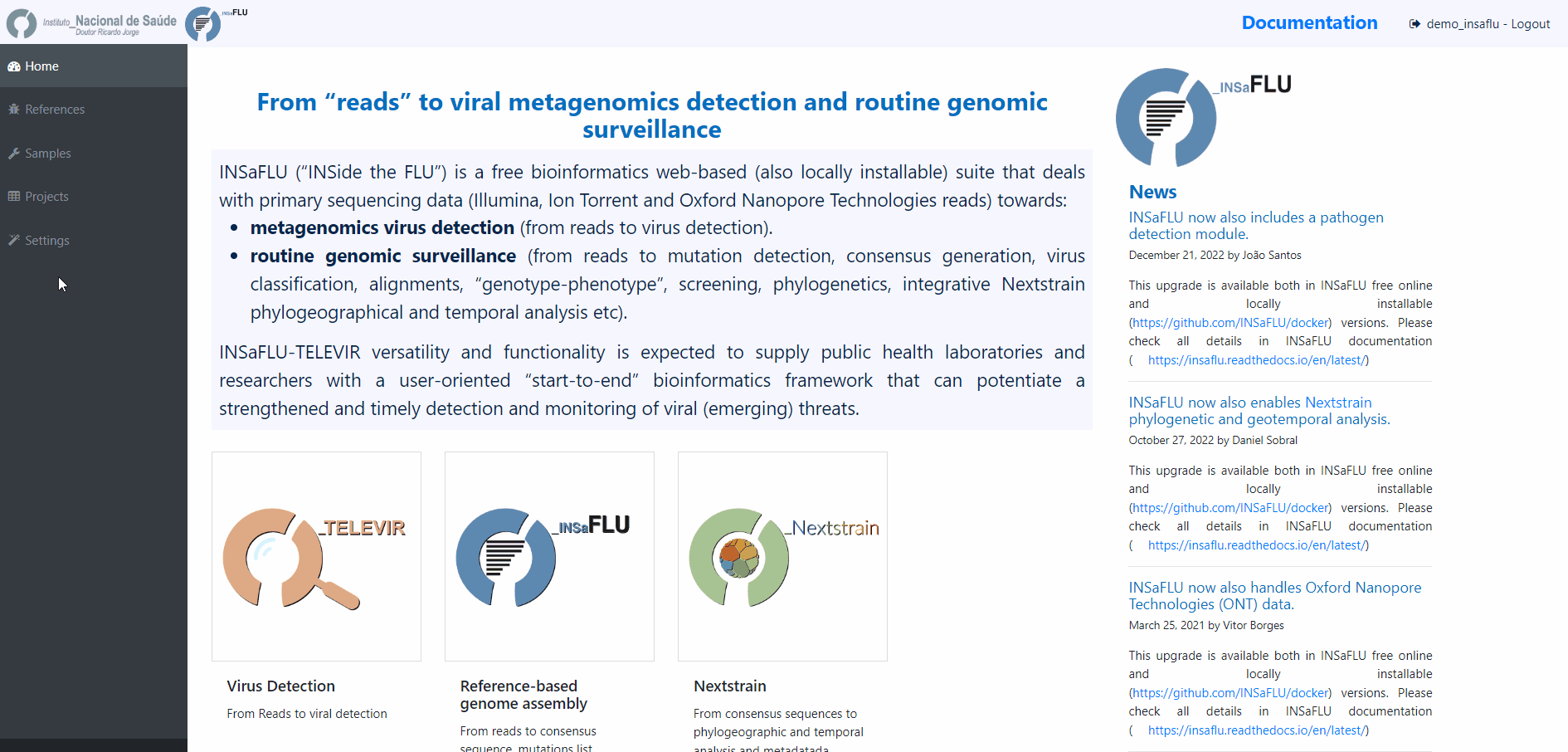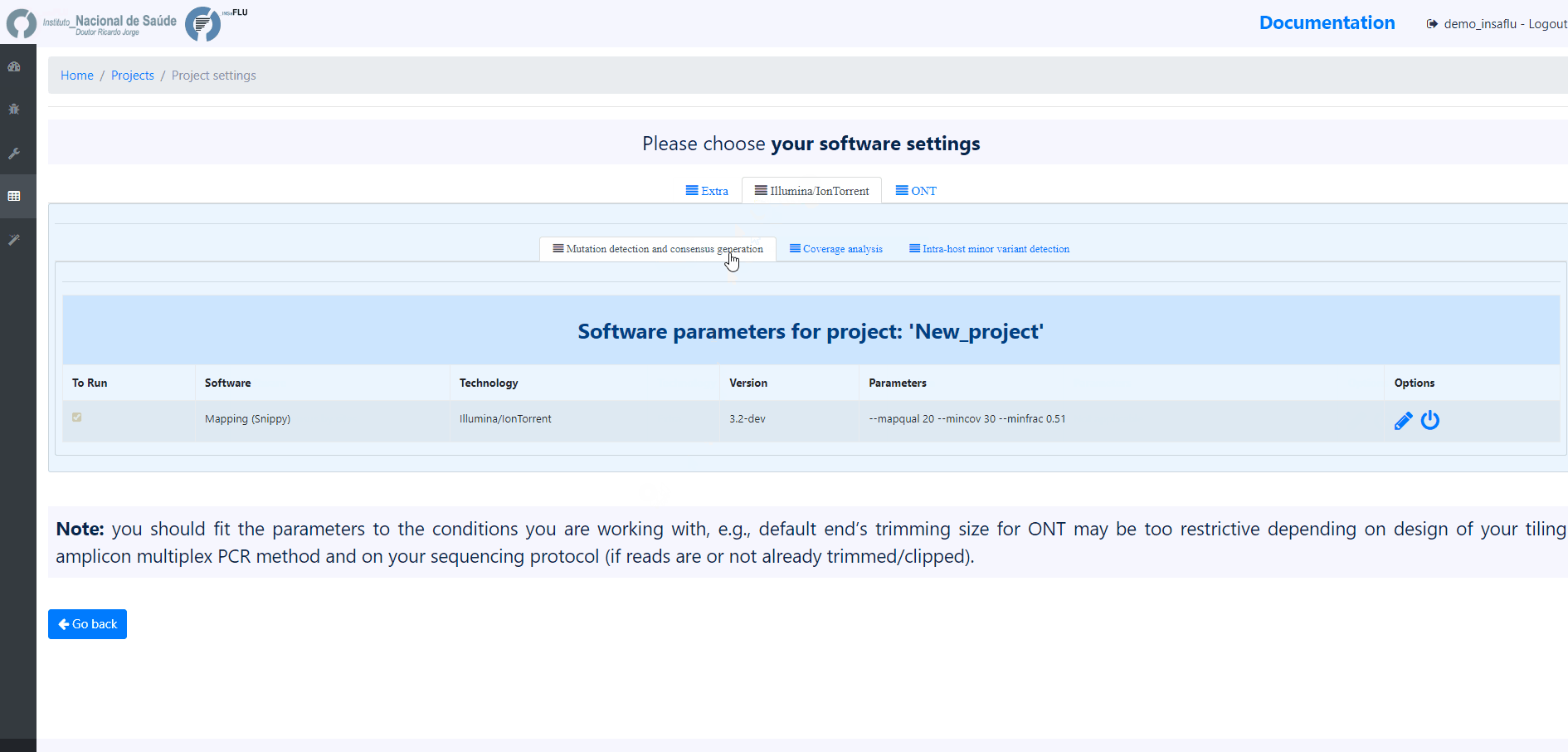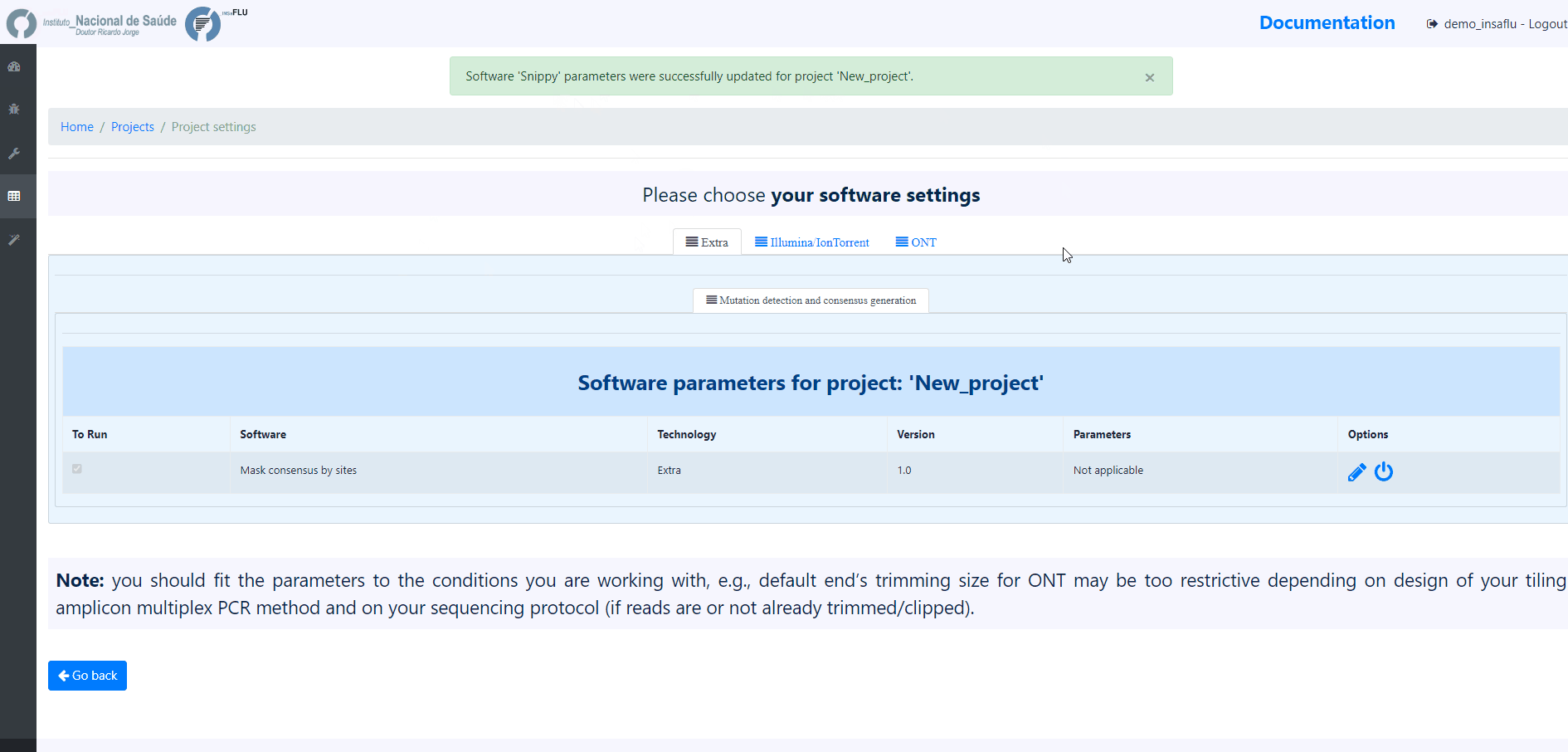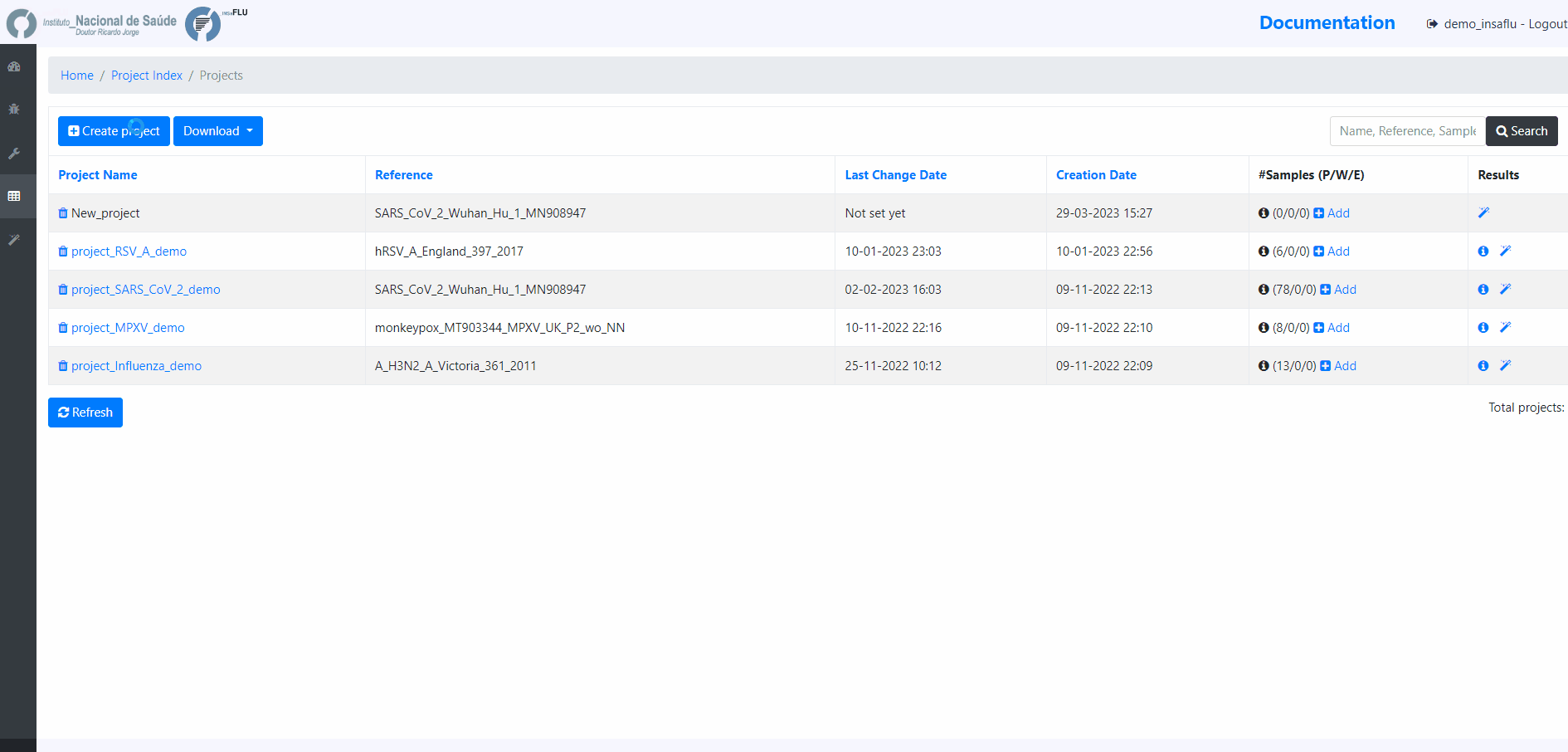
Note: Please set the parameters before assigning the first sample to the project. After that, you are still allowed to change the parameters for individual samples within the Project. Updated samples are automatically re-analysed using the novel parameters and re-inserted in the Project.
4. Add the samples to be included in the project
Example - Add a few samples
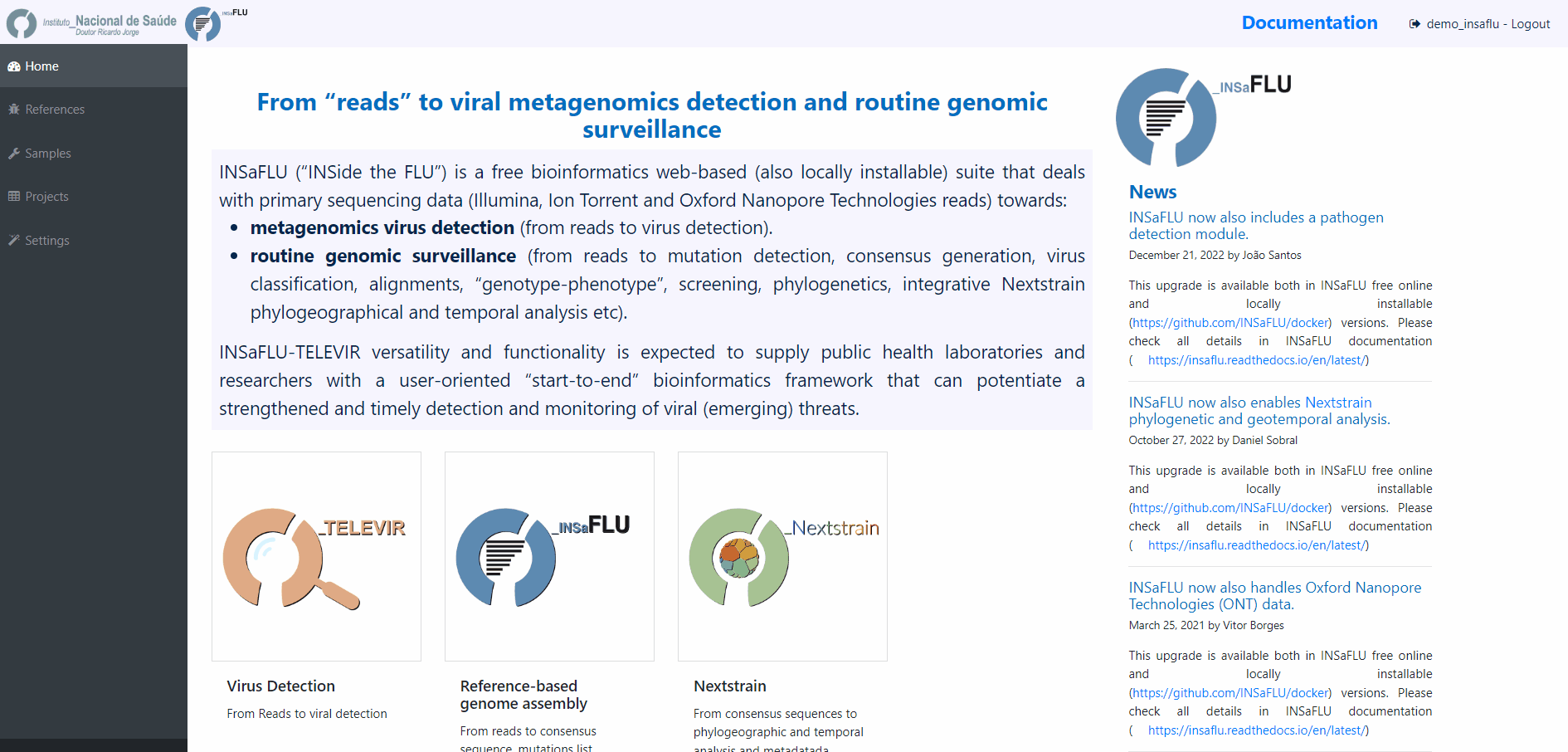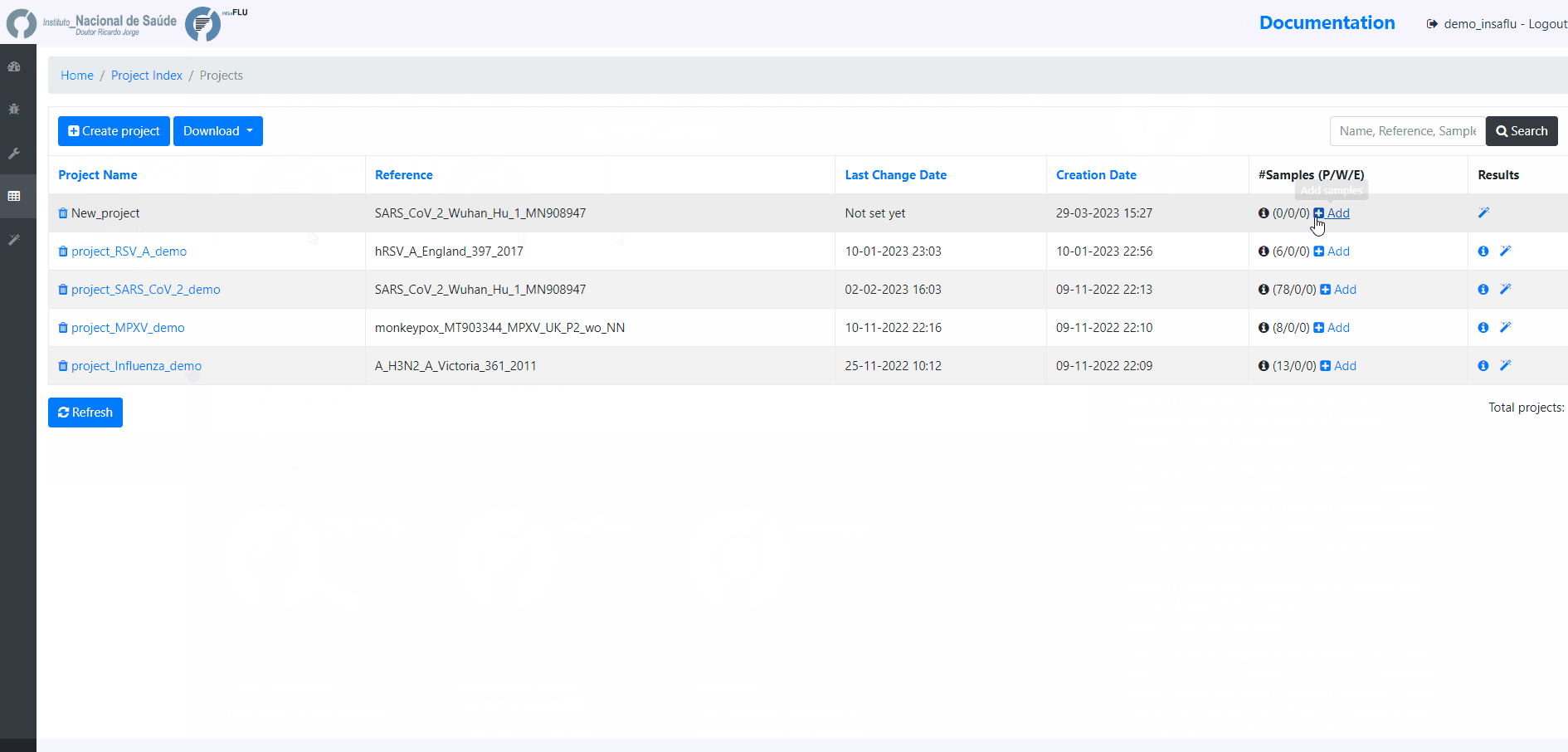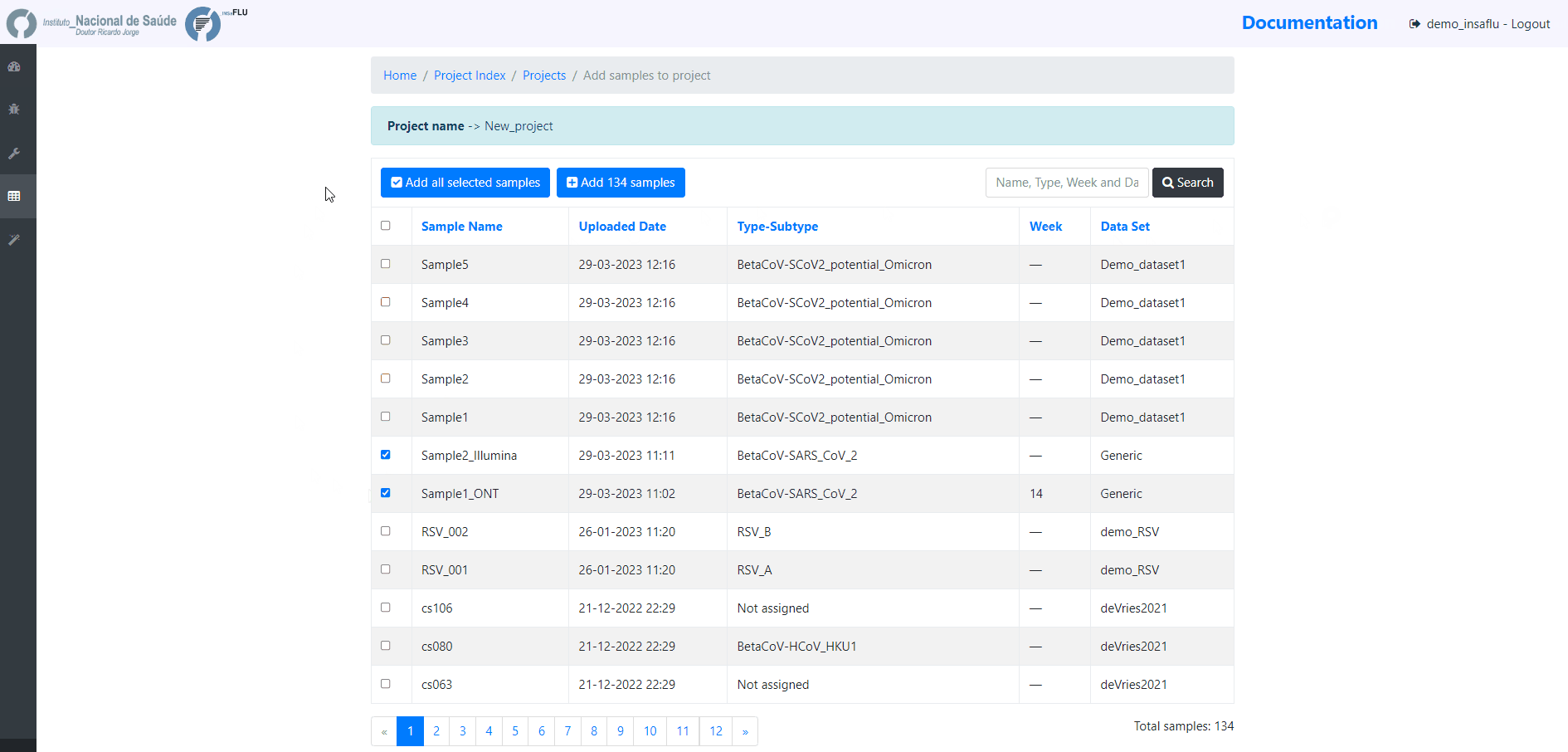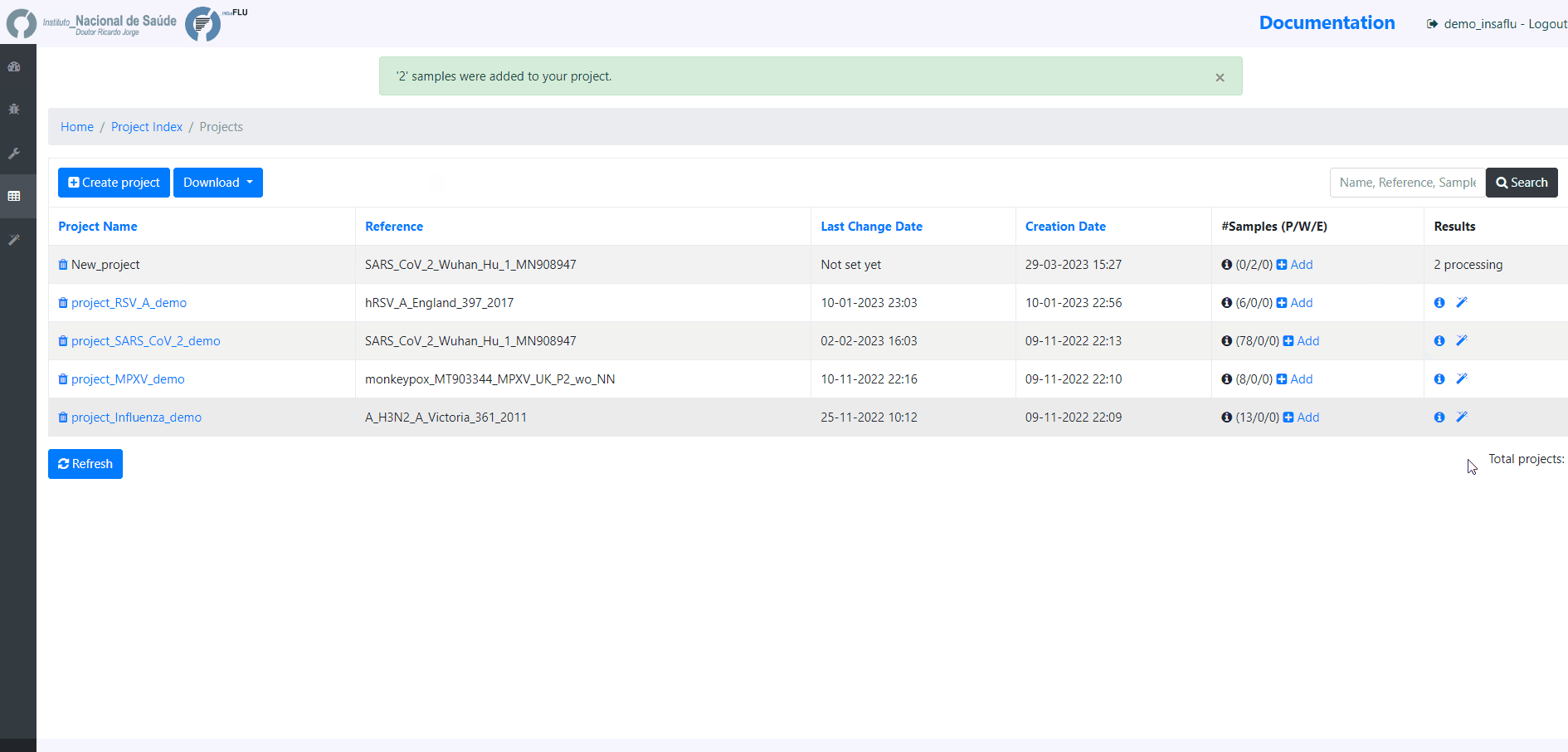
Example - Add a batch of samples (dataset)
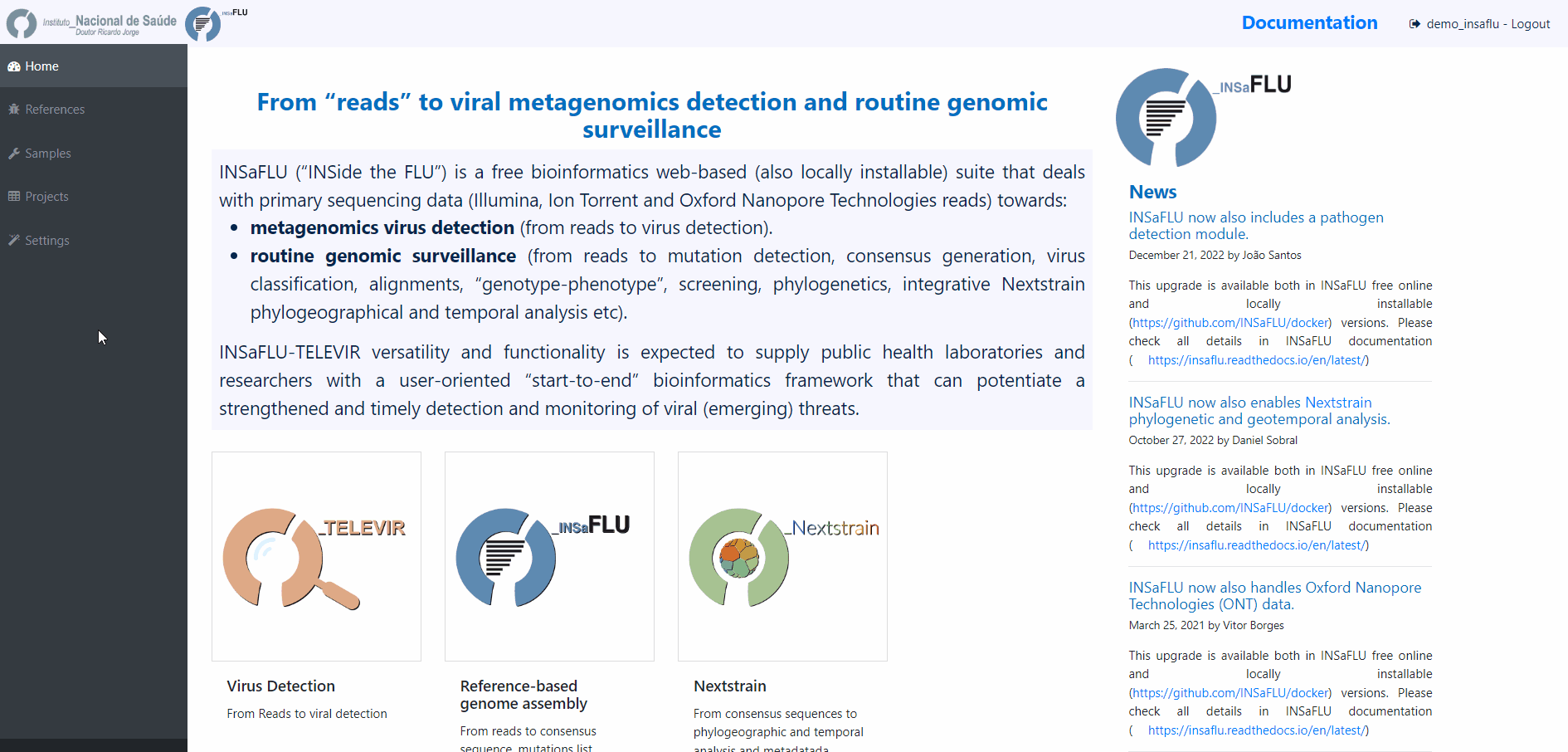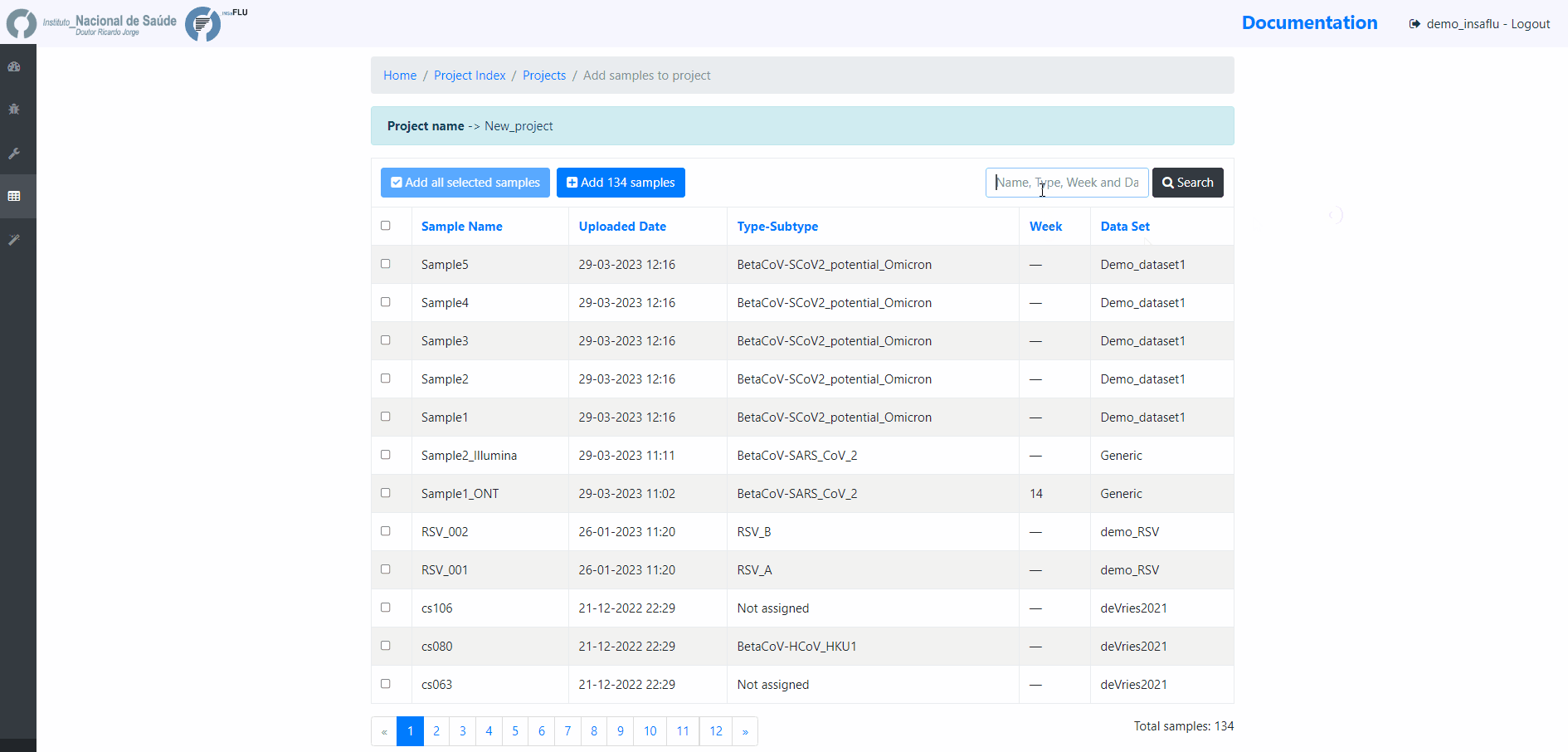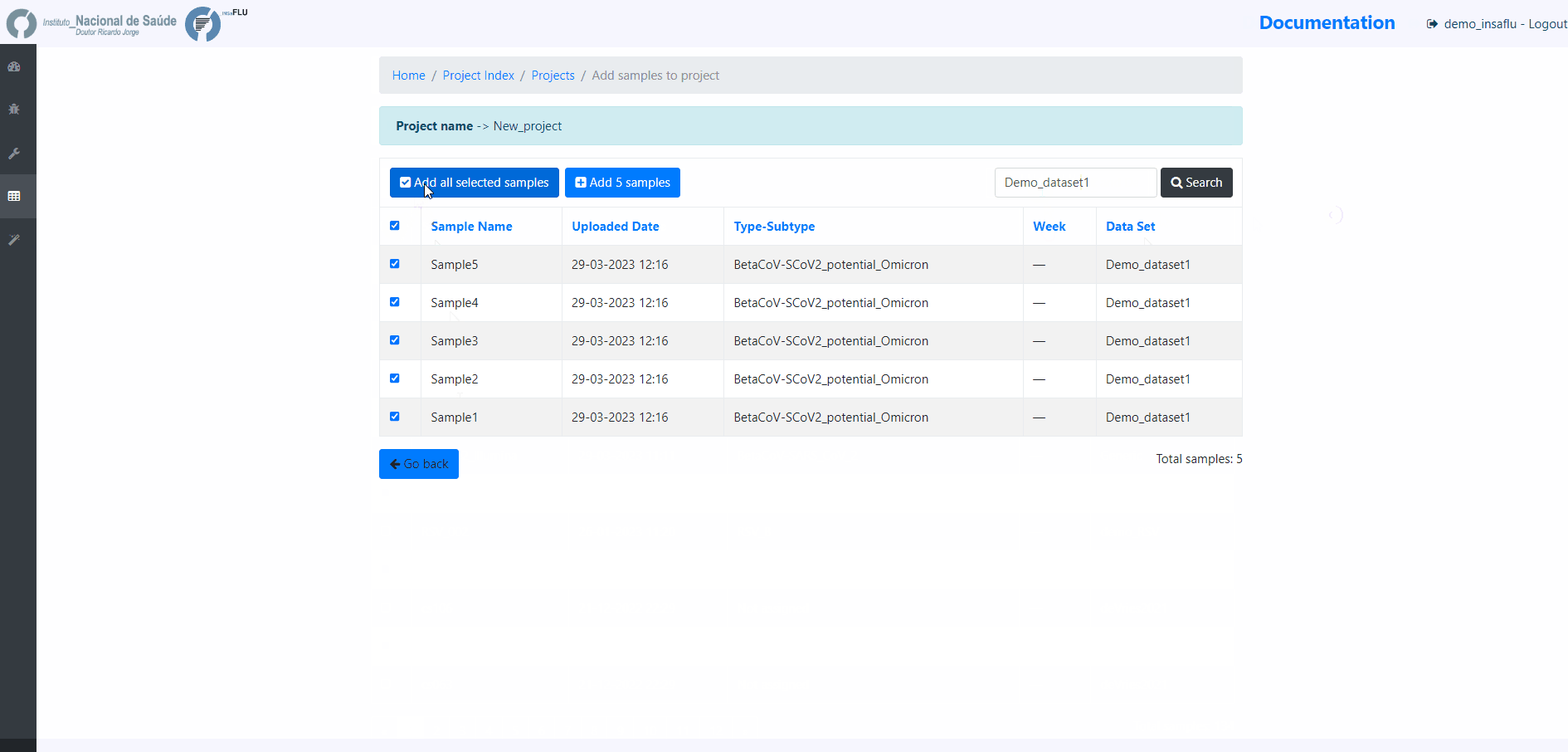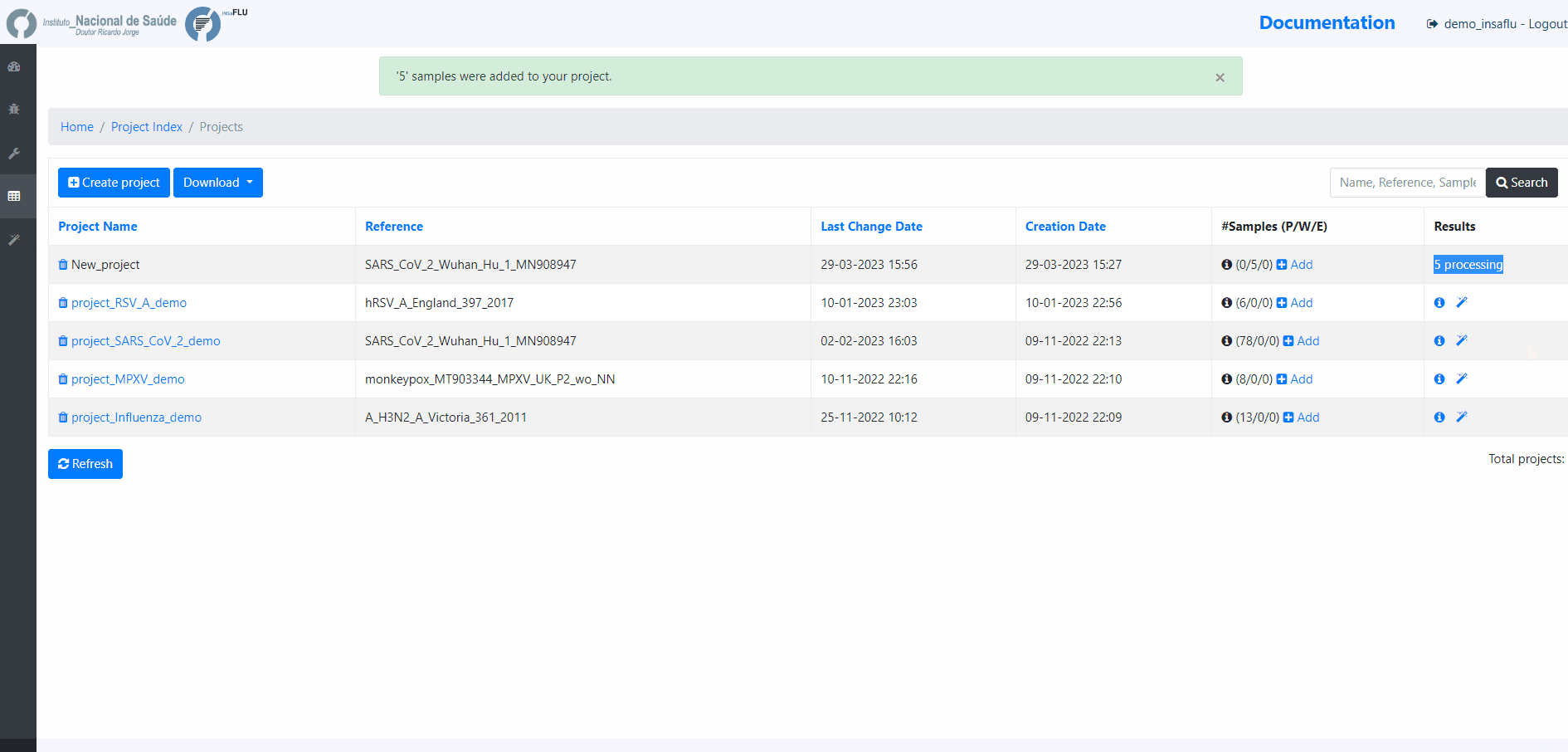
5. Monitoring Projects’ progress
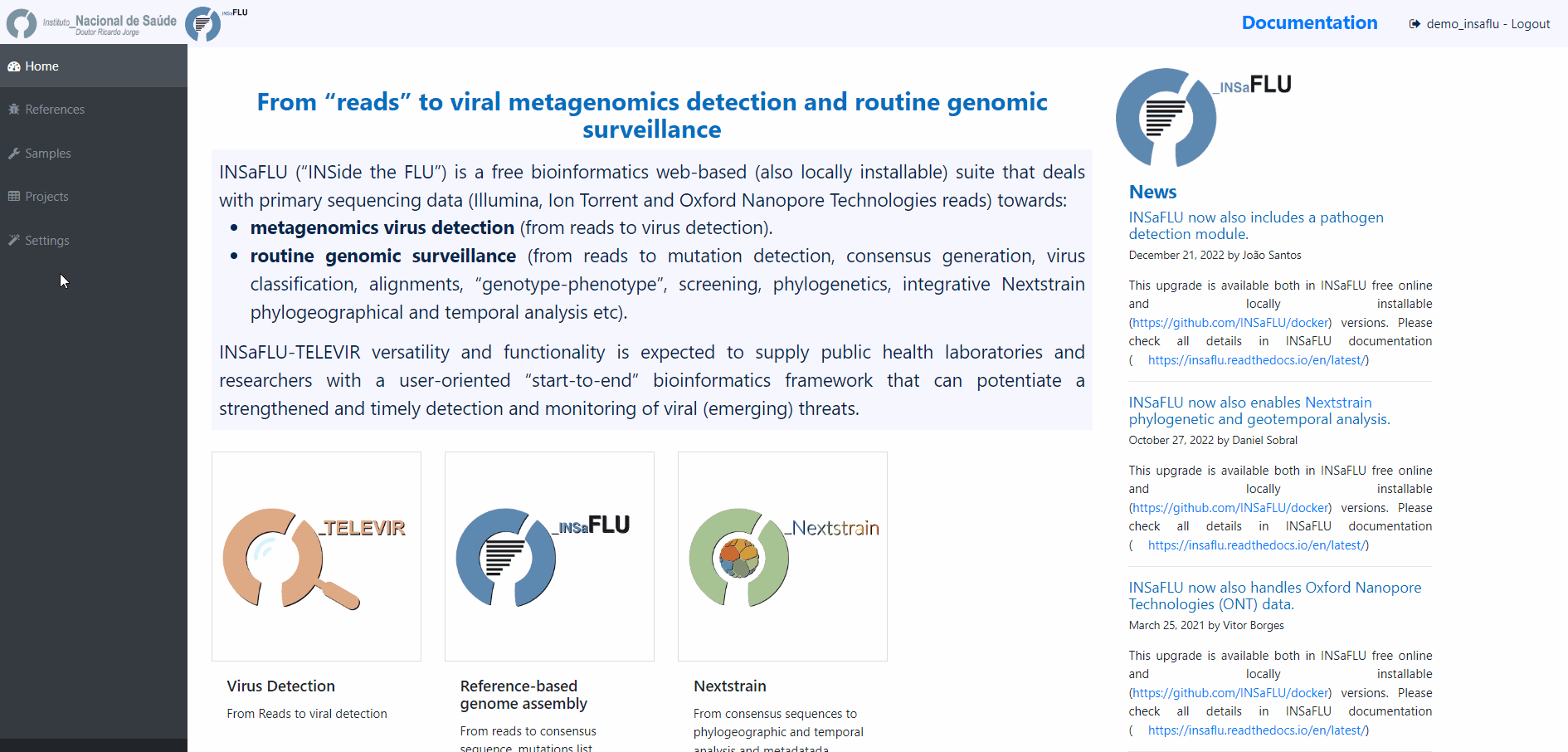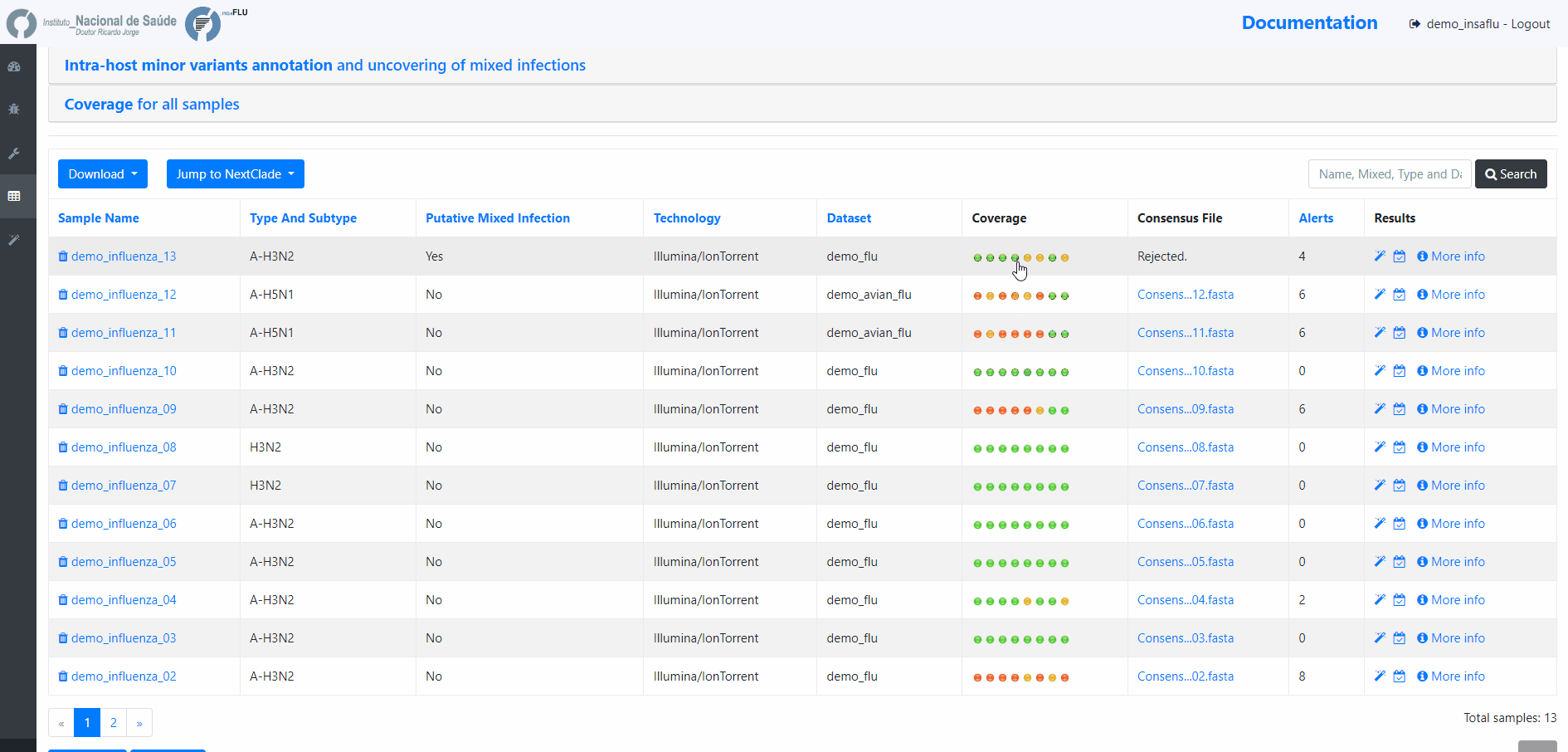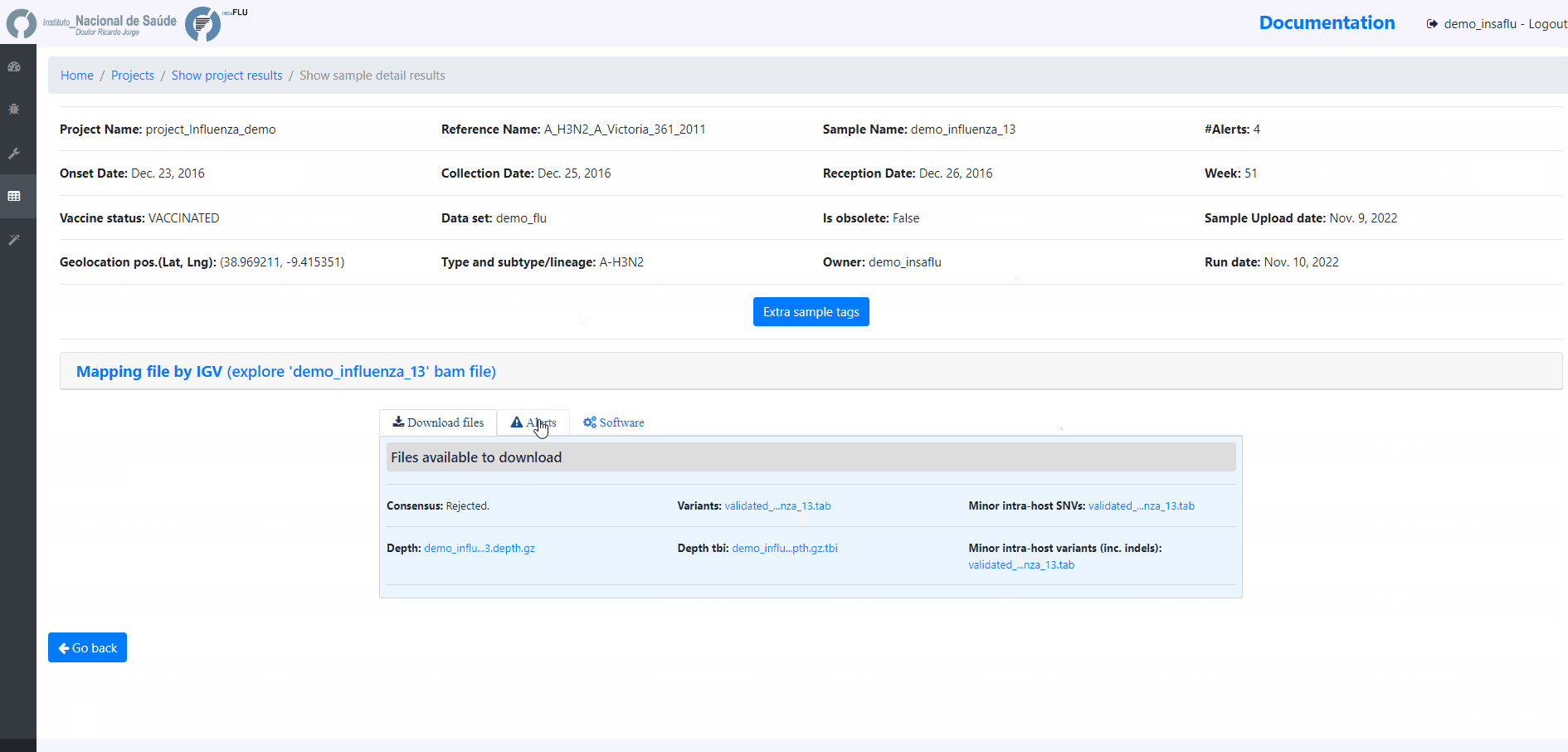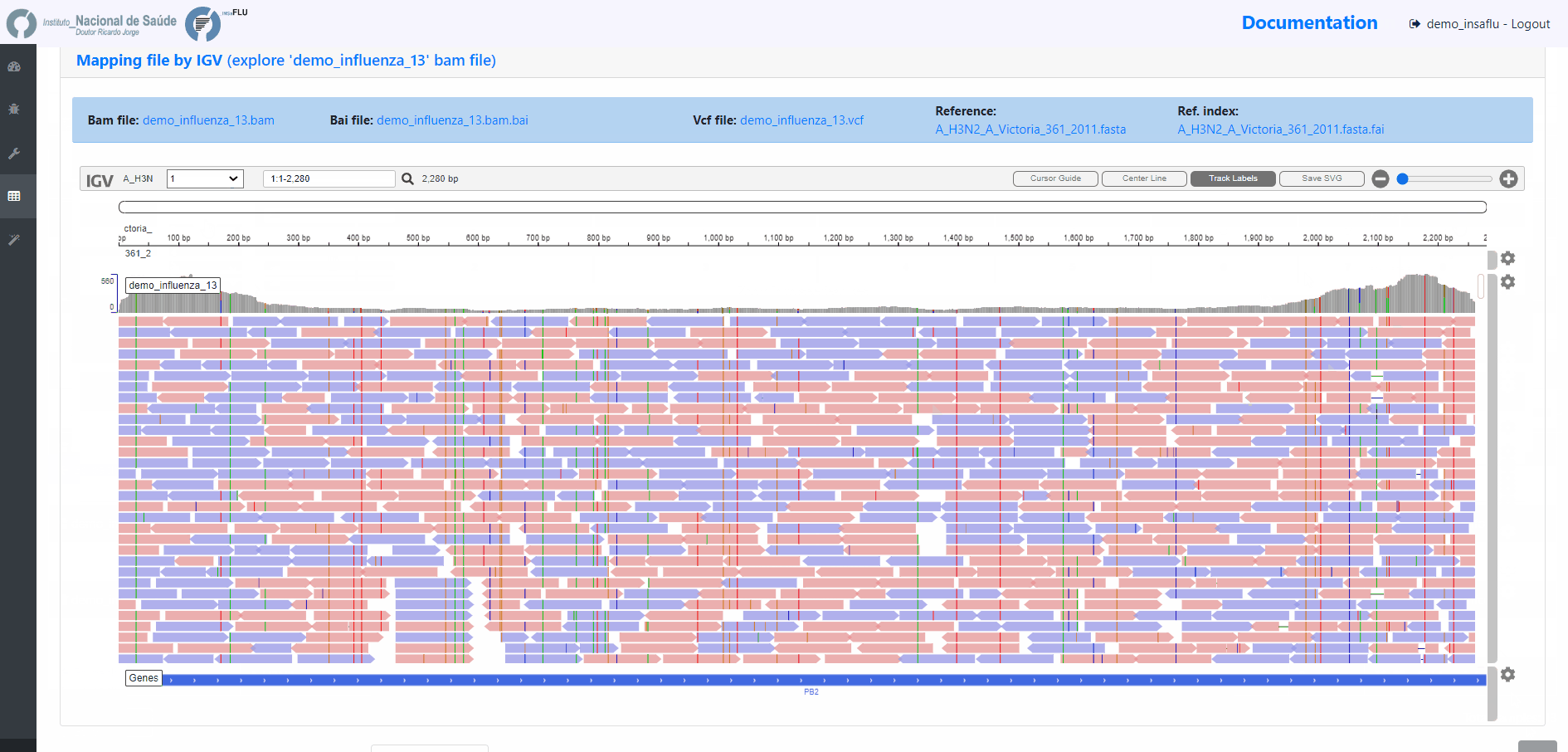
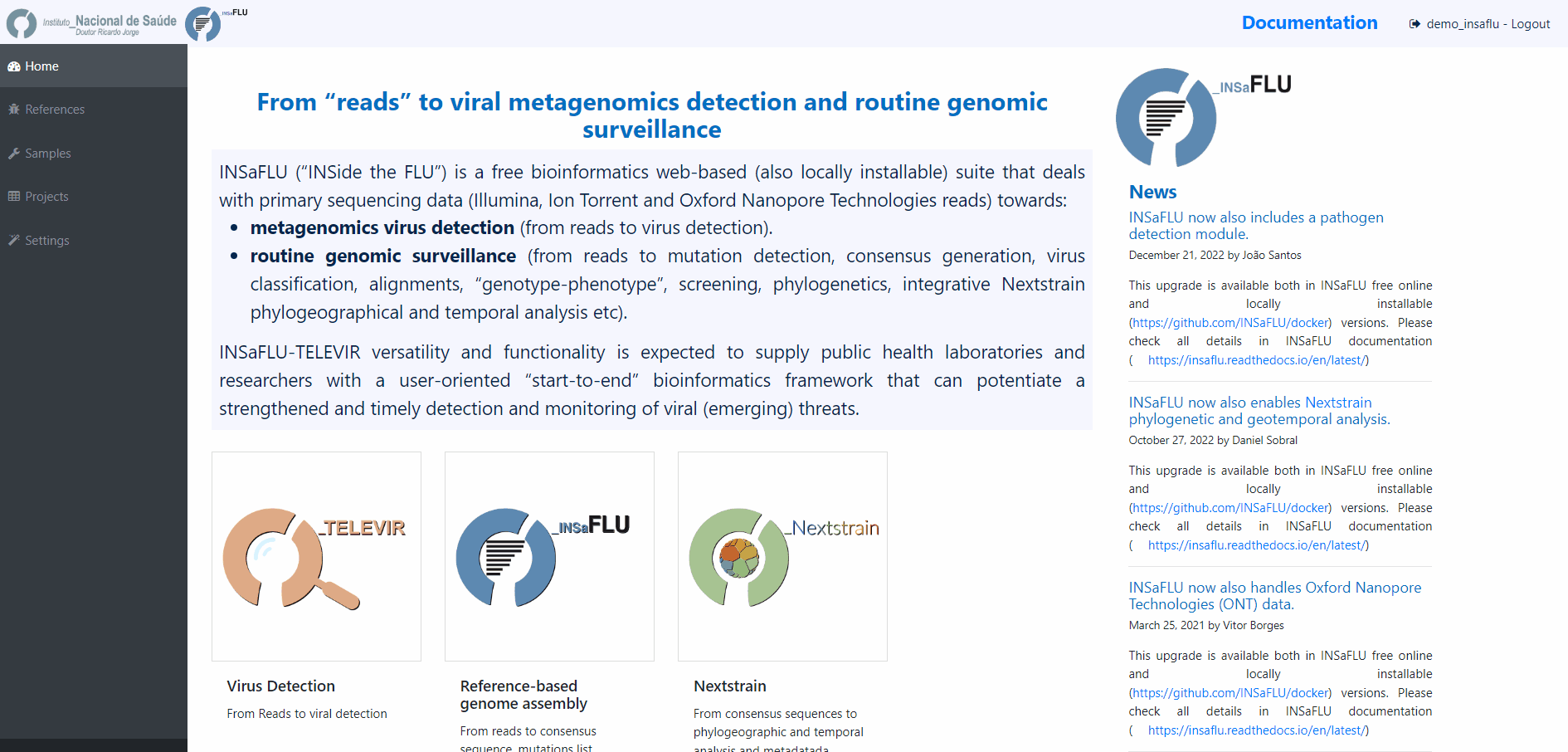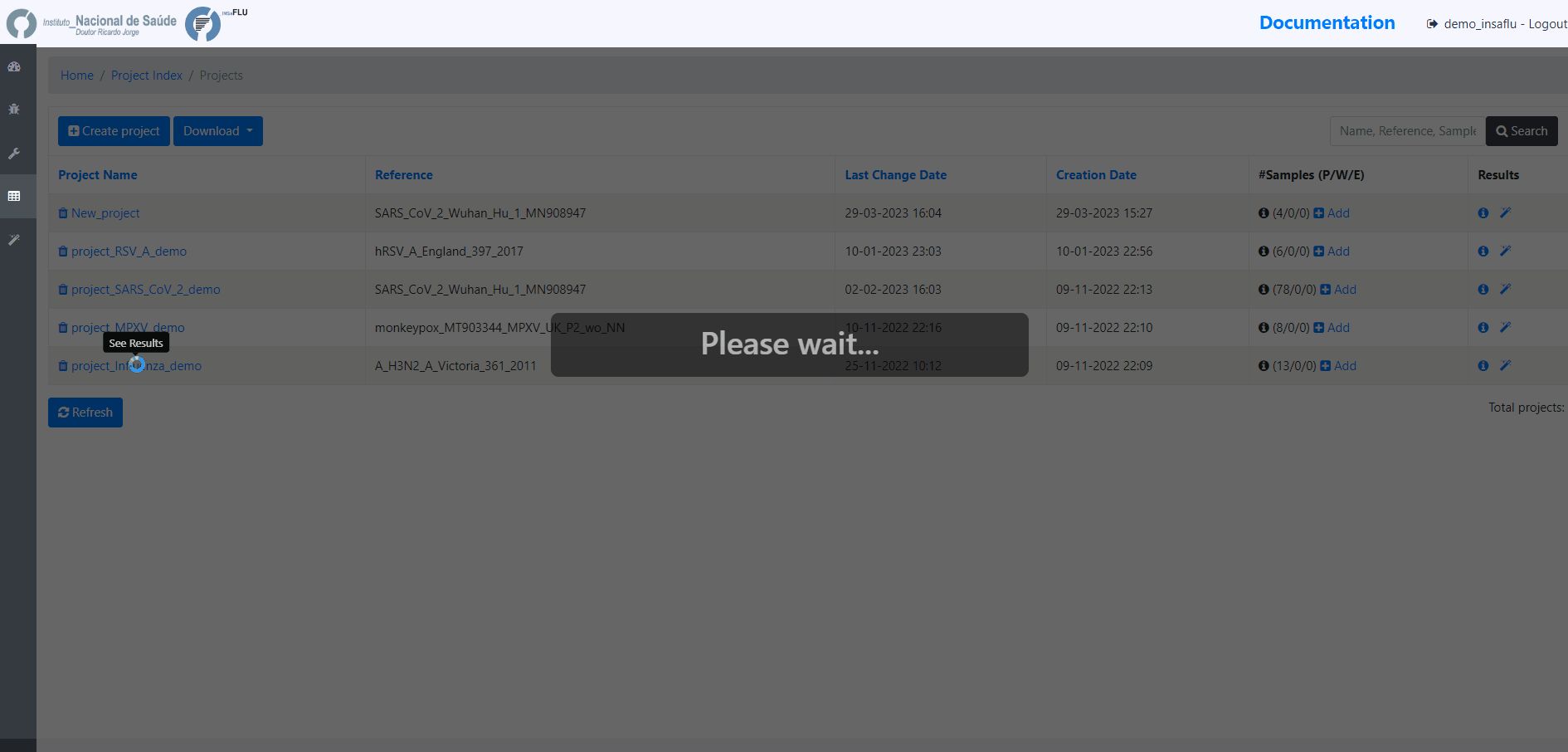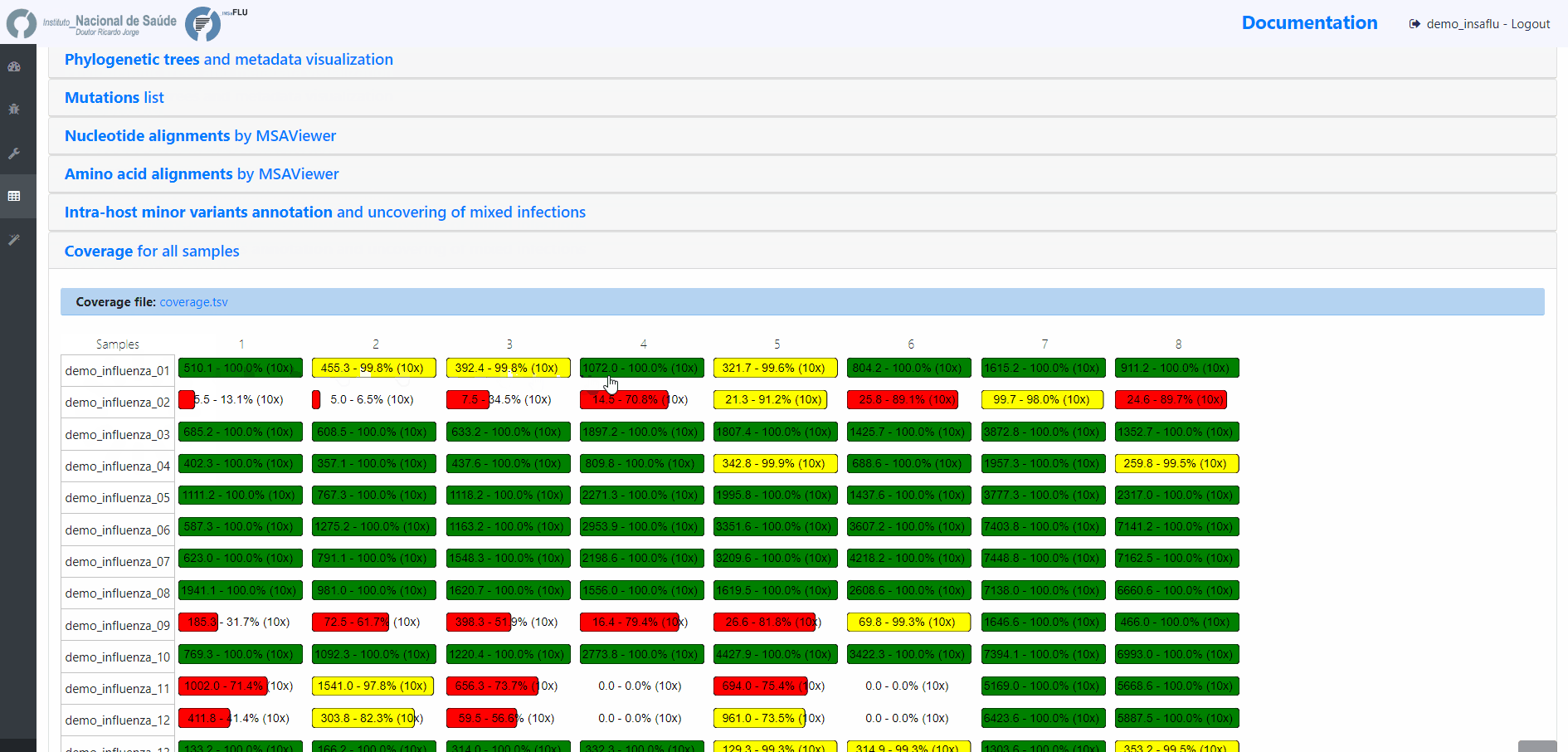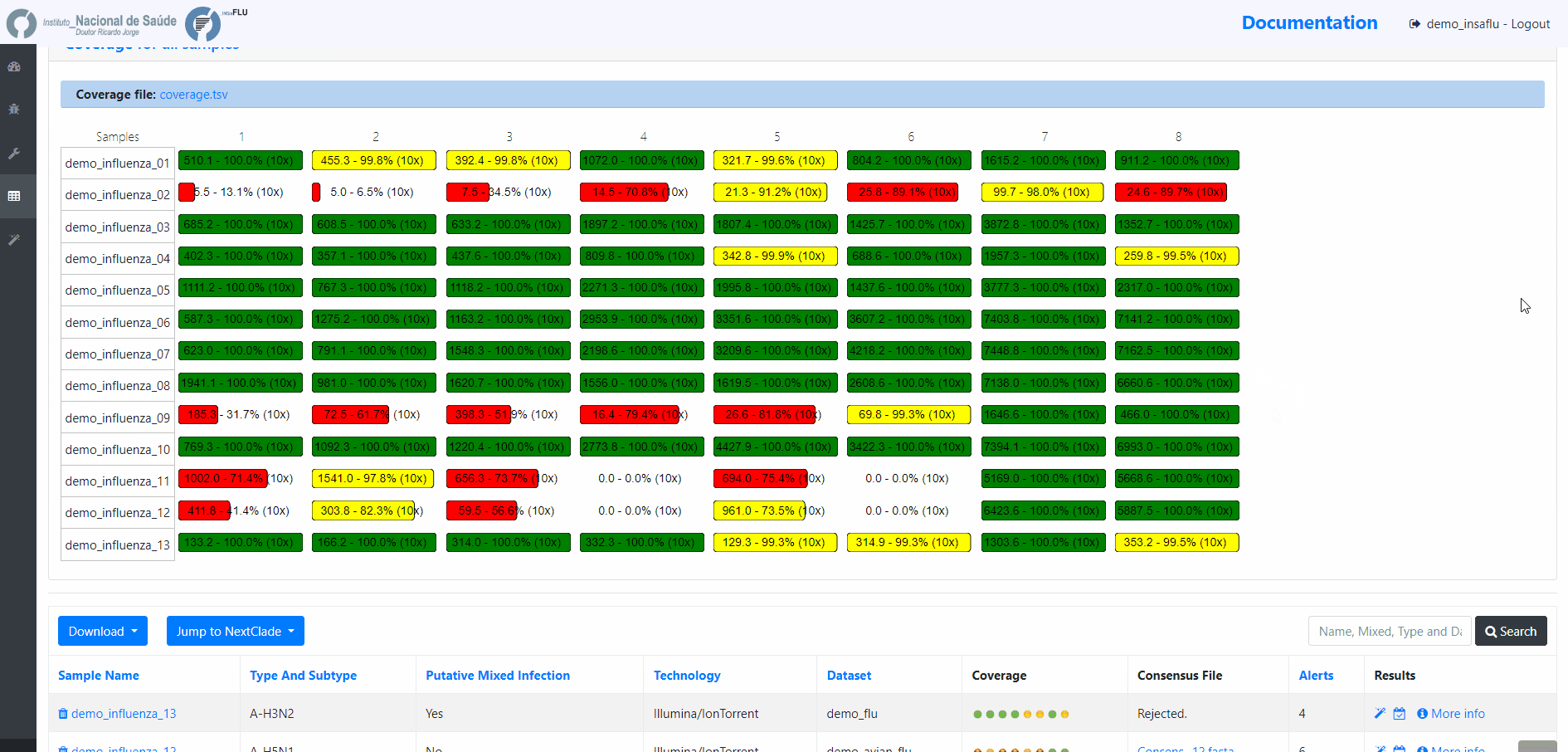
INSaFLU projects are automatically run upon creation. So, at this time, users may start monitoring the Project progress by checking the number of samples in the following status: Processed (P); Waiting (W) and Error (E).
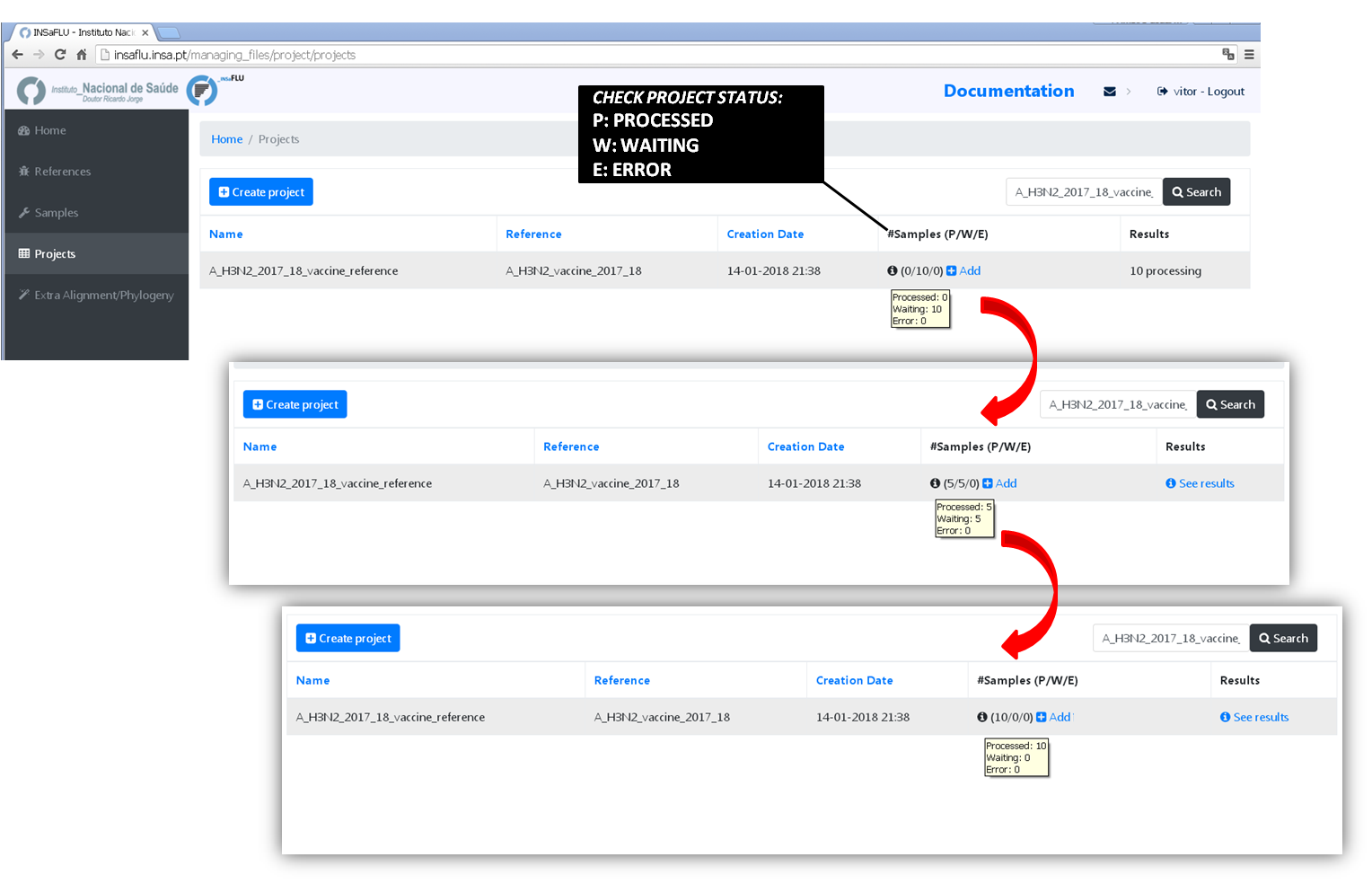
6. Scale-up your project.
You may add more samples to your Project project at any time.
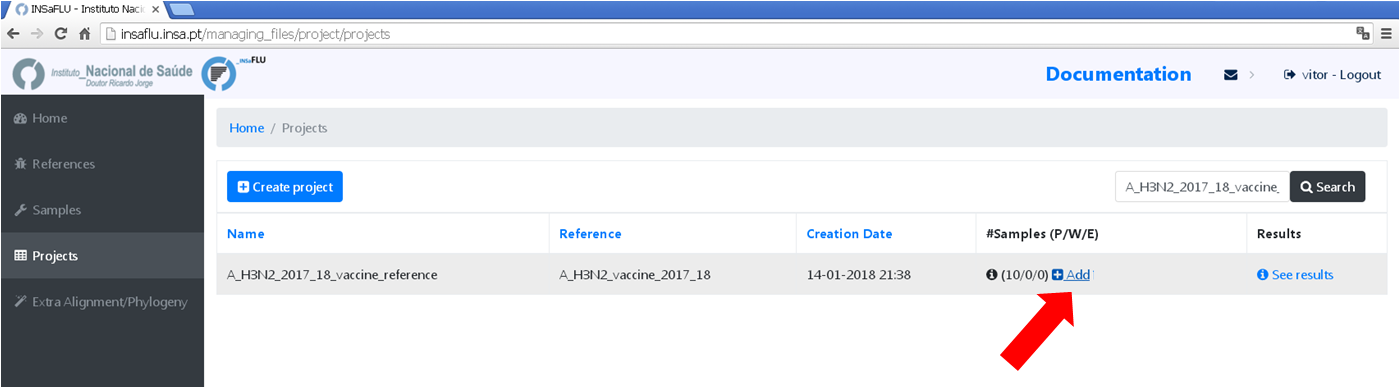
7. Modify software parameters for a given sample within a Project
Users can change the mapping parameters for individual samples within a Project. The sample is automatically re-analysed using the novel parameters and re-inserted in the Project (outputs are automatically re-calculated to integrate the “updated” sample). For instance, if the updated sample fulfill the criteria for consensus generation with the novel settings, it will be automatically integrated in the alignments and trees.
NOTE: Users can also re-run samples (with user-selected parameters) included in projects created before the 30 Oct 2020 update (see “Change log”). The updated samples will be flagged accordingly.
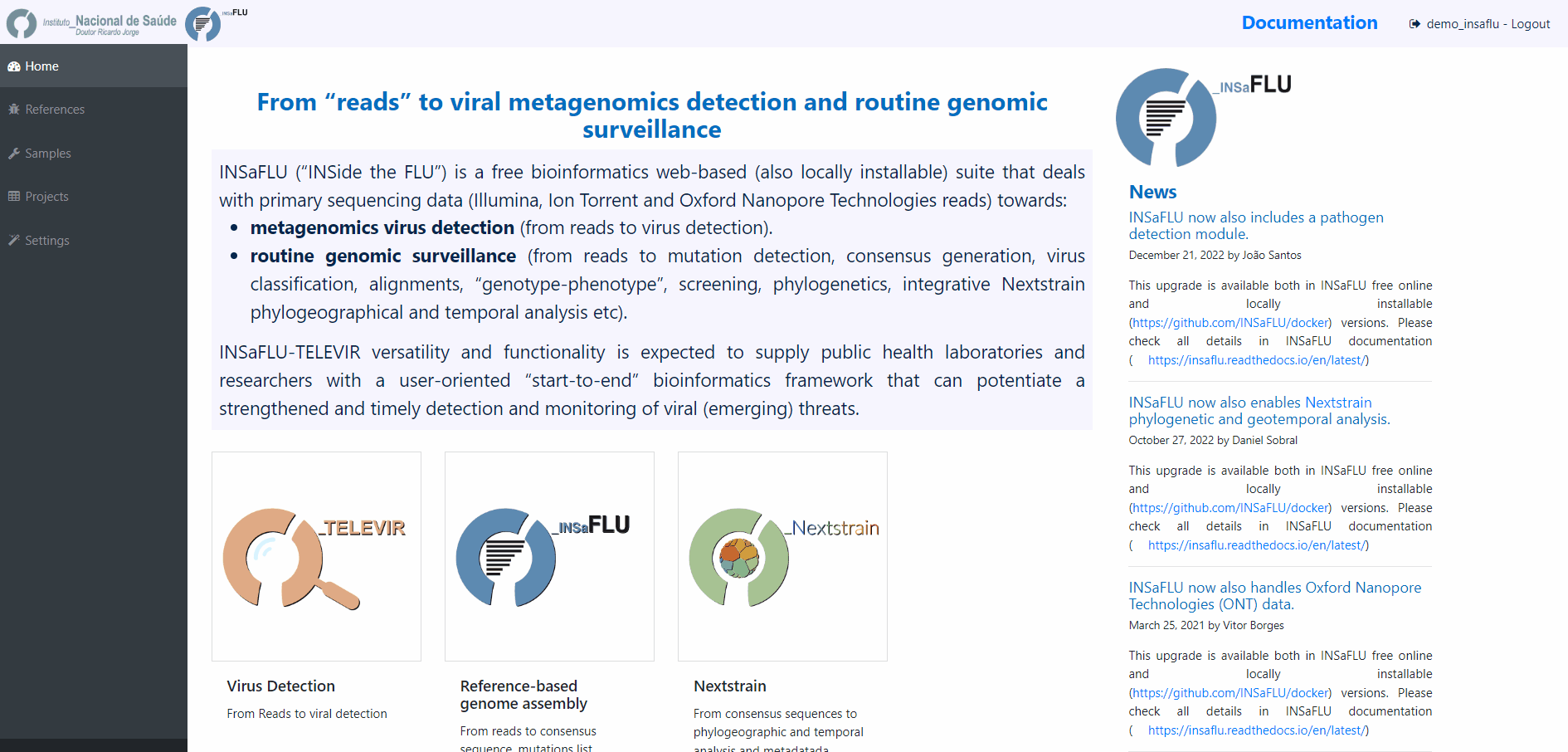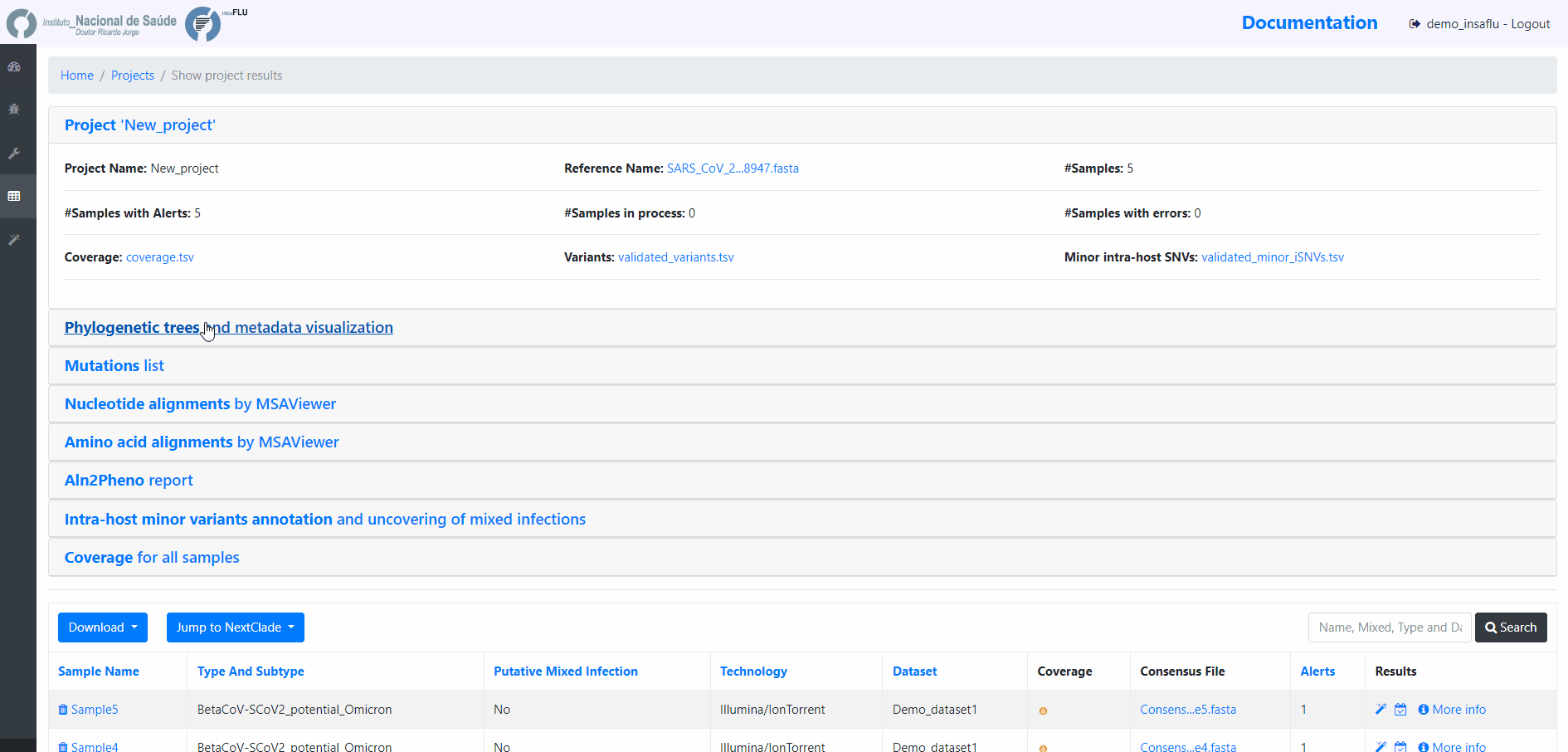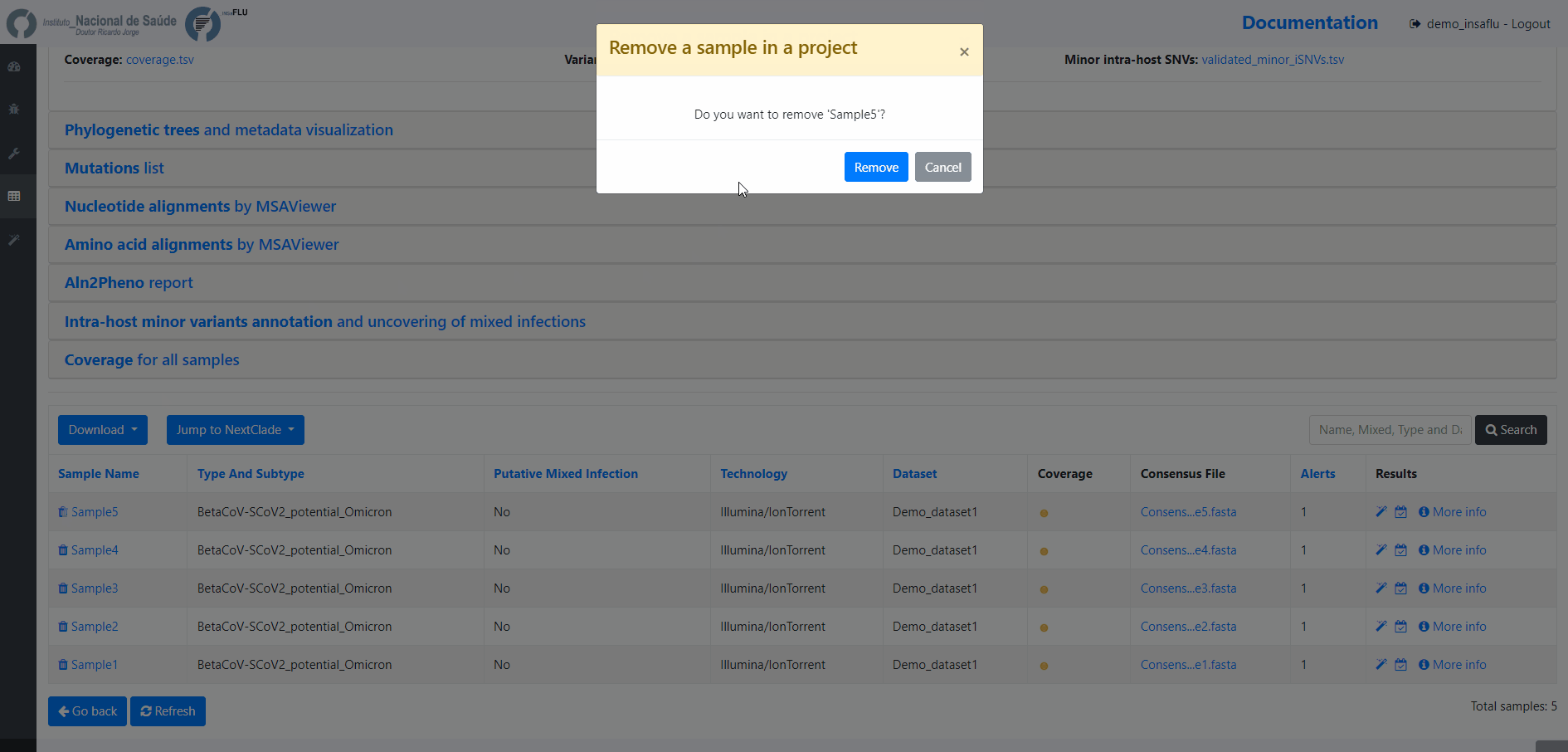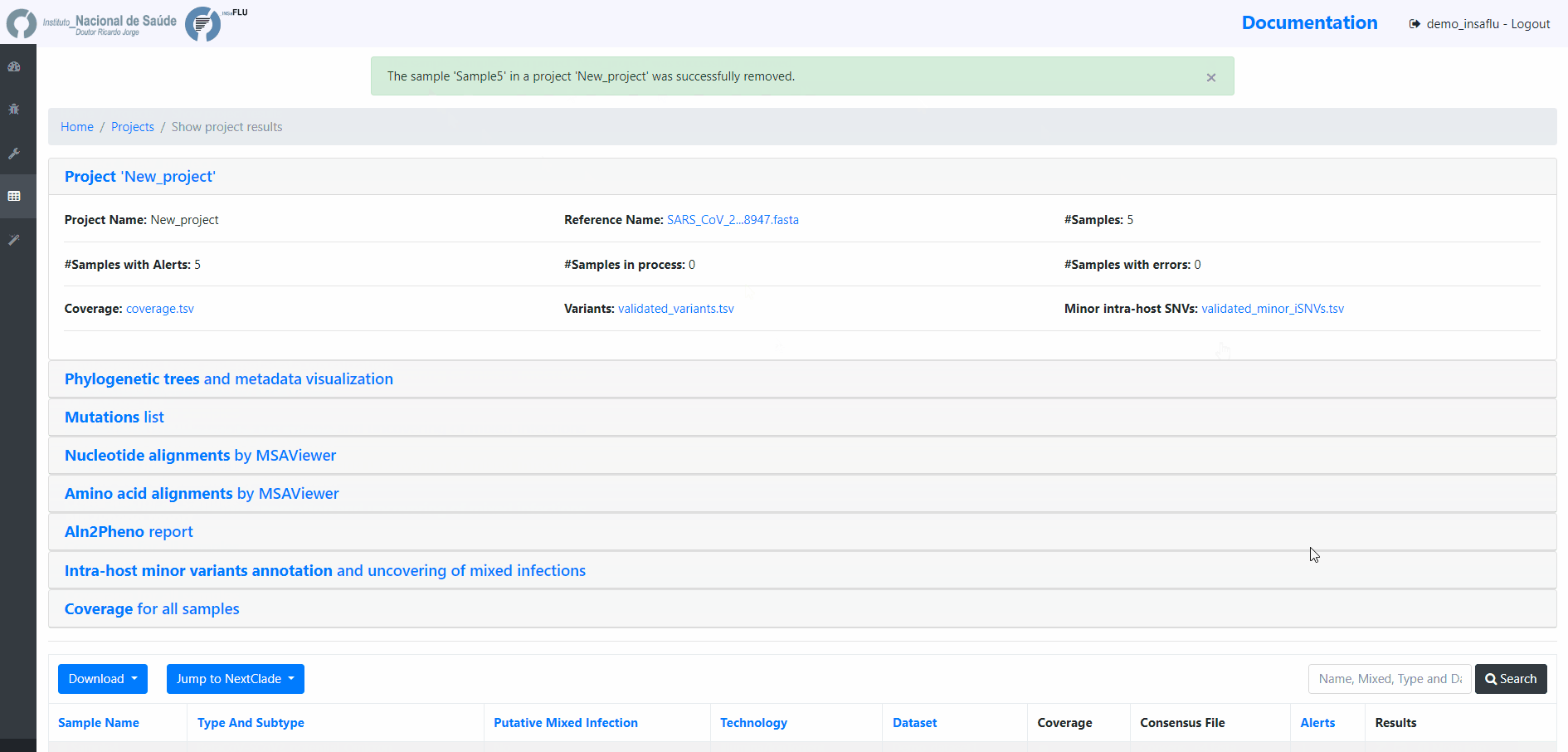
8. Remove samples from your project.
You may want to remove some samples from your project (e.g., for exclusively keeping influenza samples with success for all 8 segments)
Nextstrain Datasets - How to create and run Nextstrain datasets
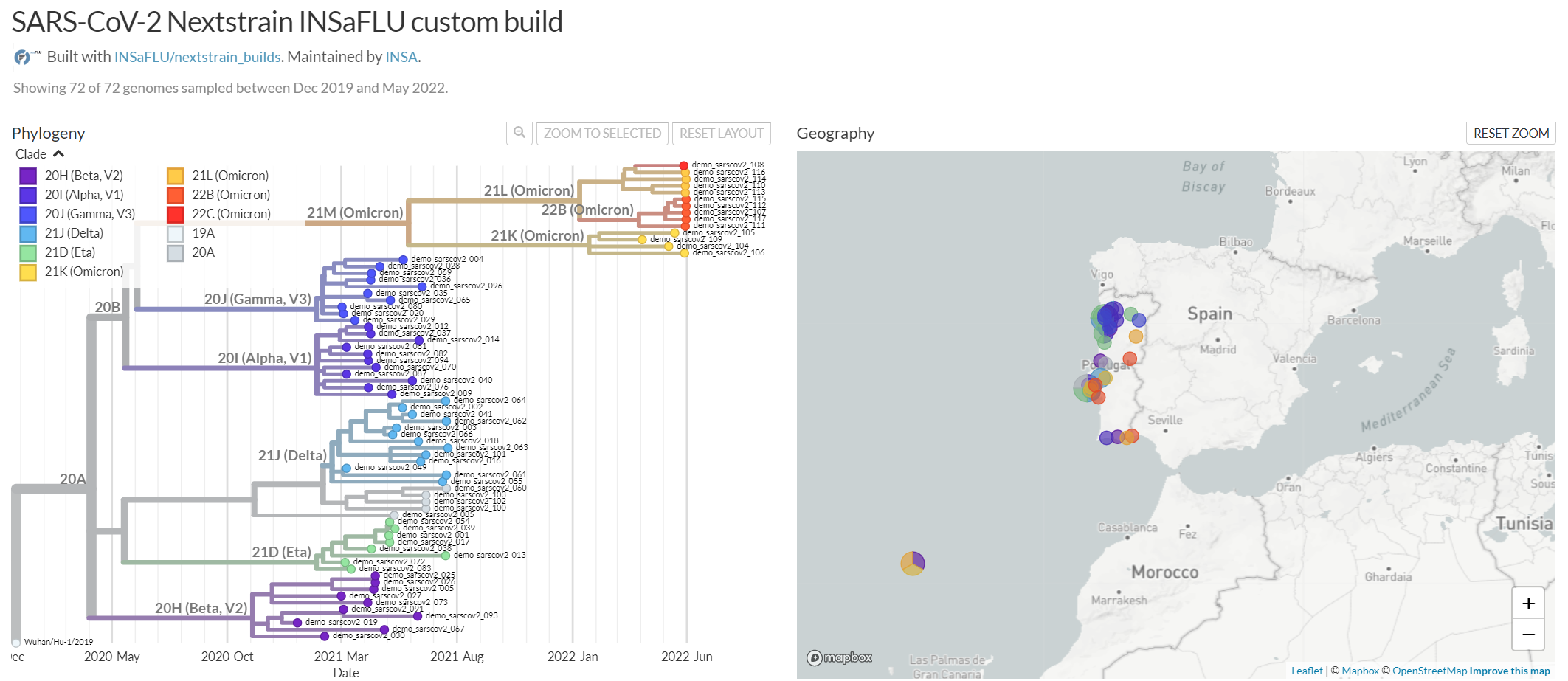
Datasets - From consensus sequences to advanced Nextstrain phylogenetic and genomic analysis, coupled with geographic and temporal data visualization and exploration of sequence metadata.
More details here: https://insaflu.readthedocs.io/en/latest/data_analysis.html#nextstrain-datasets and https://github.com/INSaFLU/nextstrain_builds
Within the Datasets menu:
2. Choose a Dataset Name and Nextstrain build and Save
[Note: the following video displays a previous version where nextstrain build was chosen after dataset creation. Now, the build is chosen when the dataset is created.]
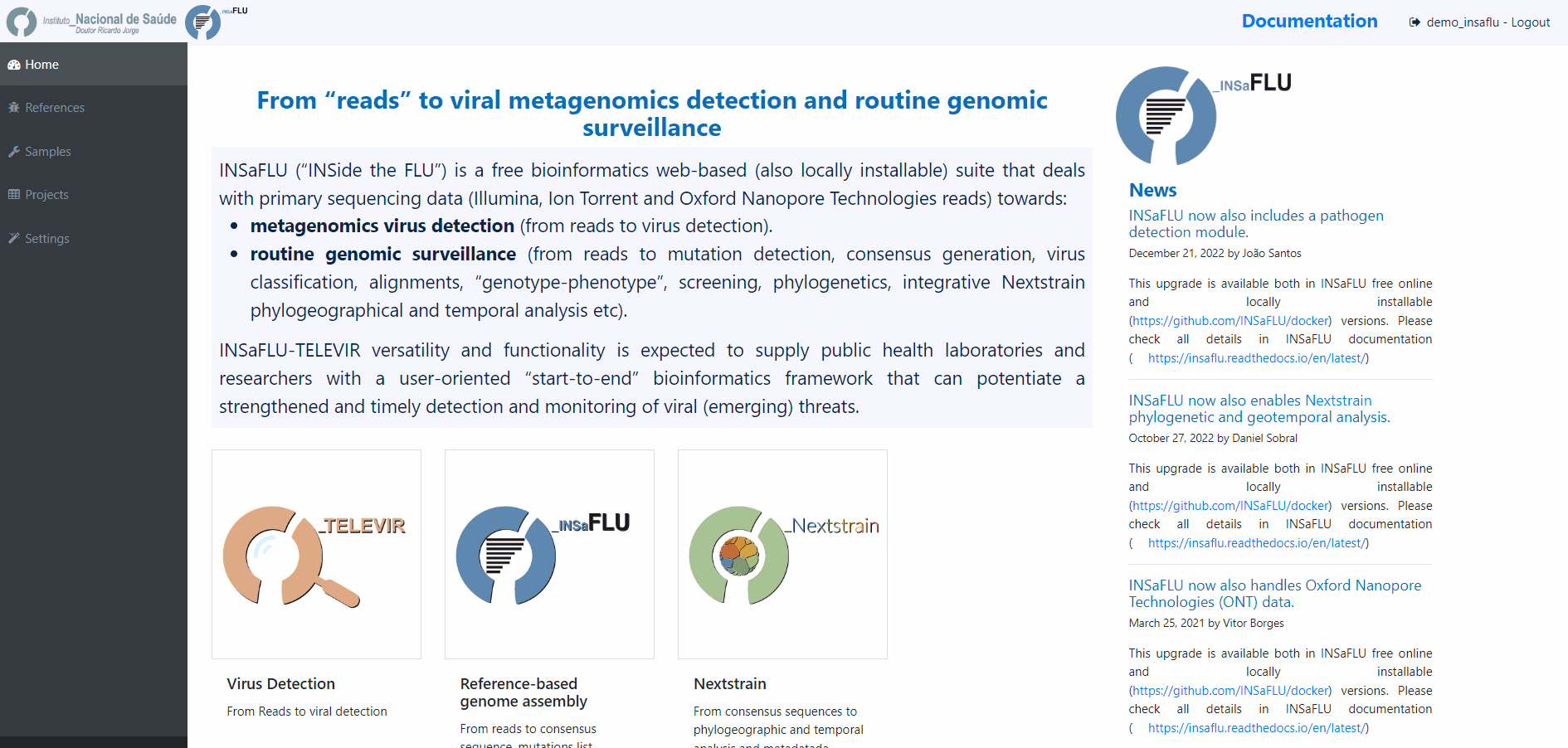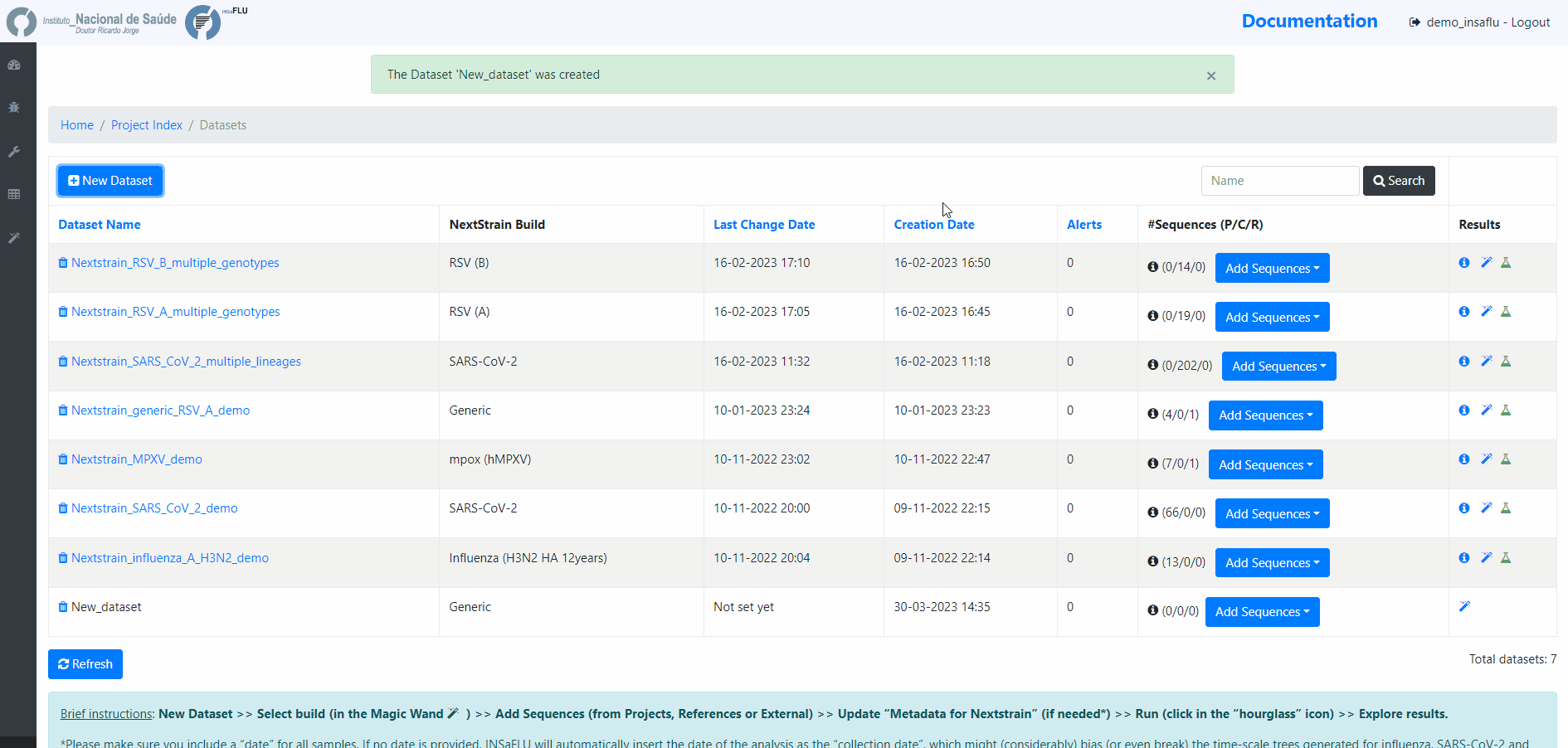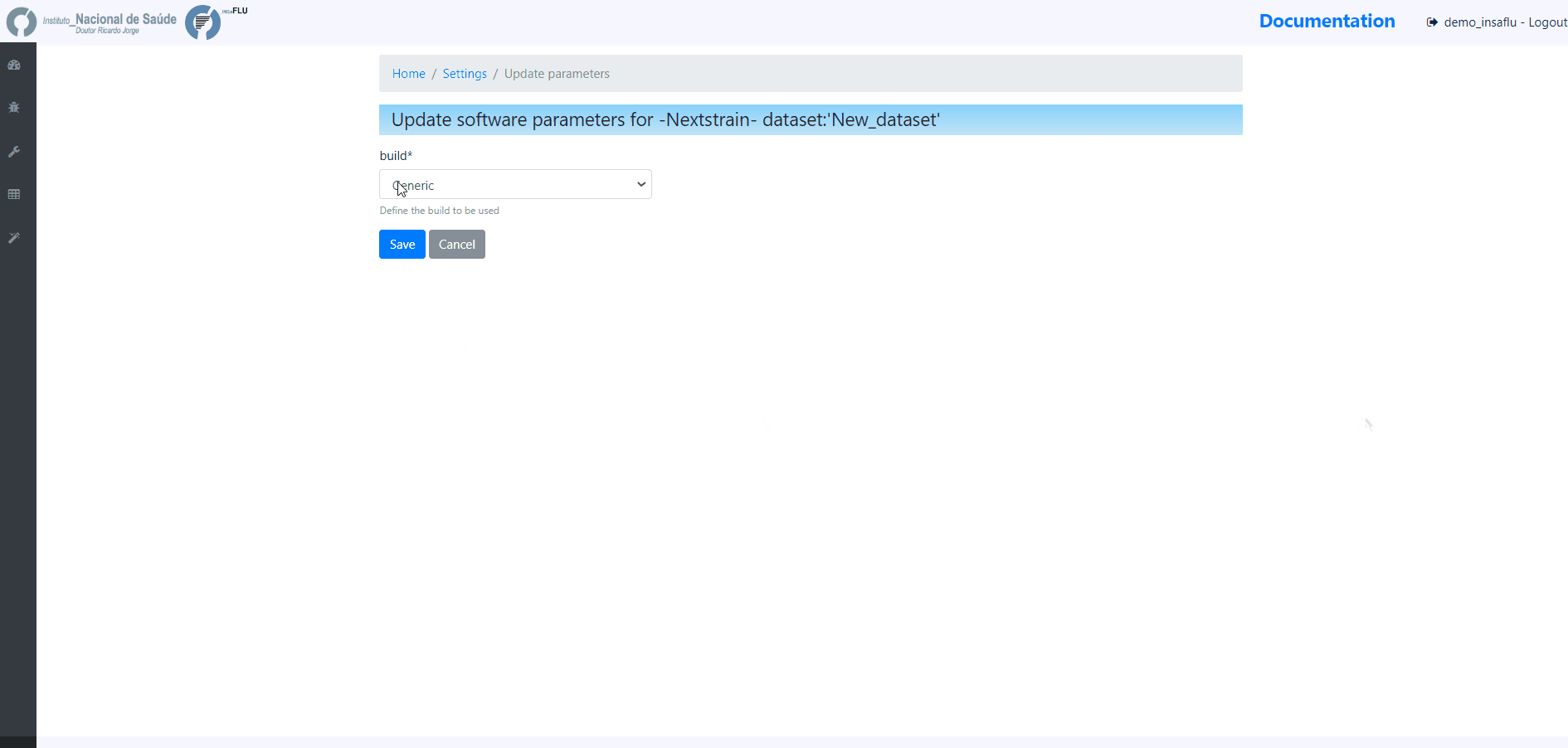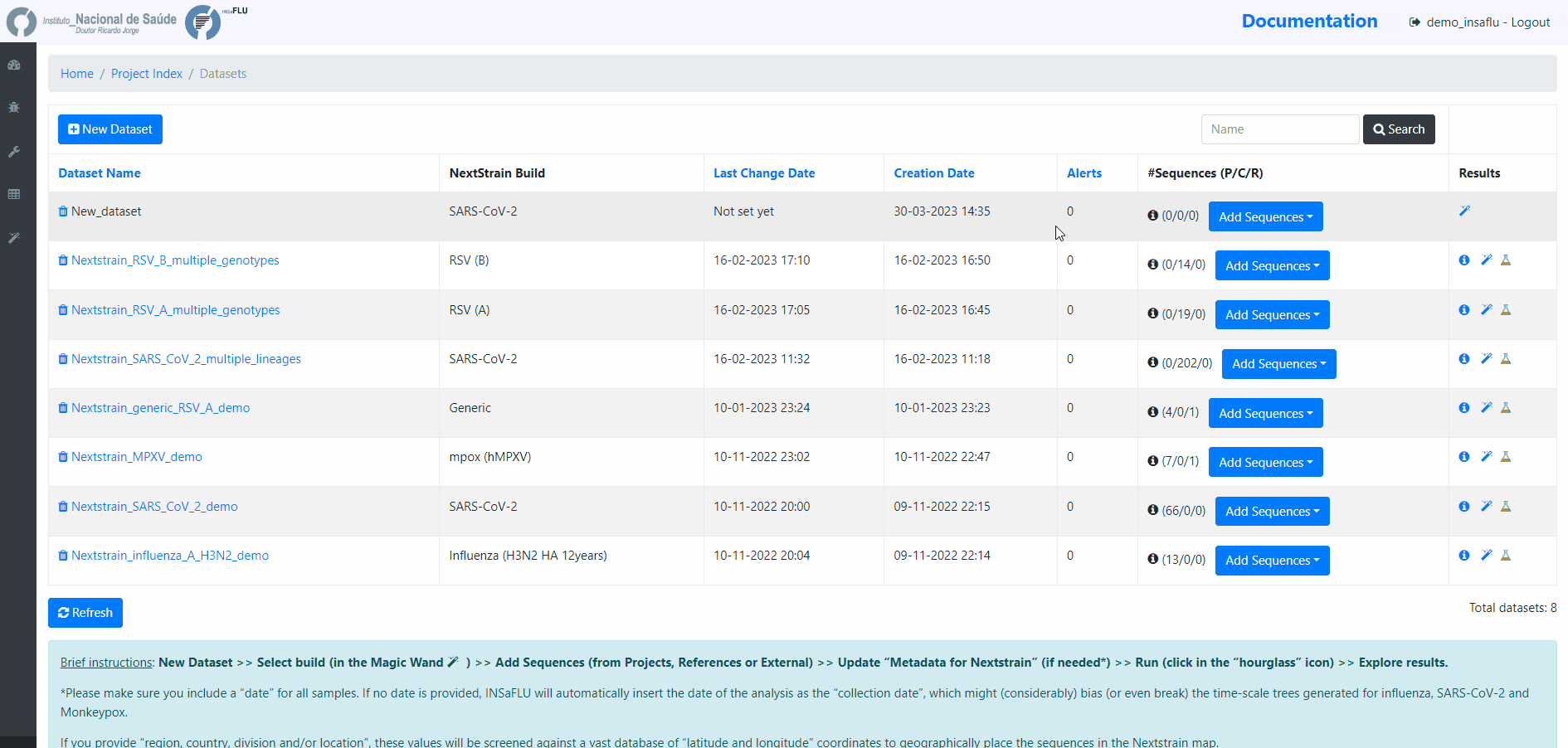
INSaFLU allows launching virus-specific Nextstrain builds (seasonal Influenza, SARS-CoV-2 and Monkeypox) as well as a “generic” build that can be used for any pathogen.
More details here: https://github.com/INSaFLU/nextstrain_builds
Builds
Seasonal influenza
INSaFLU allows running four Nexstrain builds for the seasonal influenza (A/H3N2, A/H1N1/, B/Victoria and B/Yamagata), which are simplified versions of the Influenza Nextstrain builds available at https://github.com/nextstrain/seasonal-flu
So far, influenza analyses are restricted to the Hemagglutinn (HA) coding gene. The reference HA sequences used for site (nucleotide / amino acid) numbering in the output JSON files are:
H1N1PDM: A/California/07/2009(H1N1) (https://www.ncbi.nlm.nih.gov/nuccore/CY121680.1/)
H3N2: A/Beijing/32/1992 (https://www.ncbi.nlm.nih.gov/nuccore/U26830.1/)
VIC: Influenza B virus (B/Hong Kong/02/1993) (https://www.ncbi.nlm.nih.gov/nuccore/CY018813.1/)
YAM: Influenza B virus (B/Singapore/11/1994) (https://www.ncbi.nlm.nih.gov/nuccore/CY019707.1/)
Avian influenza (under construction)
INSaFLU allows running Nexstrain builds for the avian influenza (A/H5N1), which are a simplified version of the Nextstrain builds available at https://github.com/nextstrain/avian-flu
So far, Nextstrain avian influenza can be launched for the Hemagglutinn (HA), Neuraminidase (NA) and polymerase protein PB2 (PB2) coding genes. The reference sequences used for site (nucleotide / amino acid) numbering in the output JSON files are:
HA: Influenza A virus (A/Goose/Guangdong/1/96(H5N1)) hemagglutinin (HA) (https://www.ncbi.nlm.nih.gov/nuccore/AF144305.1/)
NA: Influenza A virus (A/Goose/Guangdong/1/96(H5N1)) neuraminidase (NA) (https://www.ncbi.nlm.nih.gov/nuccore/AF144304.1)
PB2: Influenza A virus (A/Goose/Guangdong/1/96(H5N1)) polymerase (PB2)(https://www.ncbi.nlm.nih.gov/nuccore/AF144300.1)
SARS-CoV-2
This build is a simplified version of the SARS-CoV-2 Nextstrain build available at https://github.com/nextstrain/ncov
The reference genome used for site (nucleotide / amino acid) numbering and genome structure in the output JSON files is:
Wuhan-Hu-1/2019 (https://www.ncbi.nlm.nih.gov/nuccore/MN908947)
Monkeypox virus
This build is a simplified version of the Monkeypox virus Nextstrain build available at https://github.com/nextstrain/monkeypox
The reference genome used for site (nucleotide / amino acid) numbering and genome structure in the output JSON files is:
MPXV-M5312_HM12_Rivers (https://www.ncbi.nlm.nih.gov/nuccore/NC_063383)
Respiratory Syncytial Virus (RSV)
This build is a simplified version of the RSV virus Nextstrain build available at https://github.com/nextstrain/rsv
The reference genomes used for site (nucleotide / amino acid) numbering and genome structure in the output JSON files is:
RSV A: A/England/397/2017 (GISAID ID EPI_ISL_412866)
RSV B: B/Australia/VIC-RCH056/2019 (GISAID ID EPI_ISL_1653999)
Generic
This build is a simplified version of the Nextstrain build available at https://github.com/nextstrain/zika
This generic build uses as reference sequence (as tree root and for mutation annotation) one of the reference sequences of the projects included in the Nextstrain dataset.
Currently, the generic build does not generate a Time-Resolved Tree (unlike the virus-specific builds).
Generic with Time Tree
This build is the same as the generic build but also runs a time tree. To make use of this build, samples need to have associated dates.
4. Add the samples to be included in the Dataset
You can add samples to the Dataset from different sources:
Projects - user-selected consensus sequences generated within INSaFLU projects
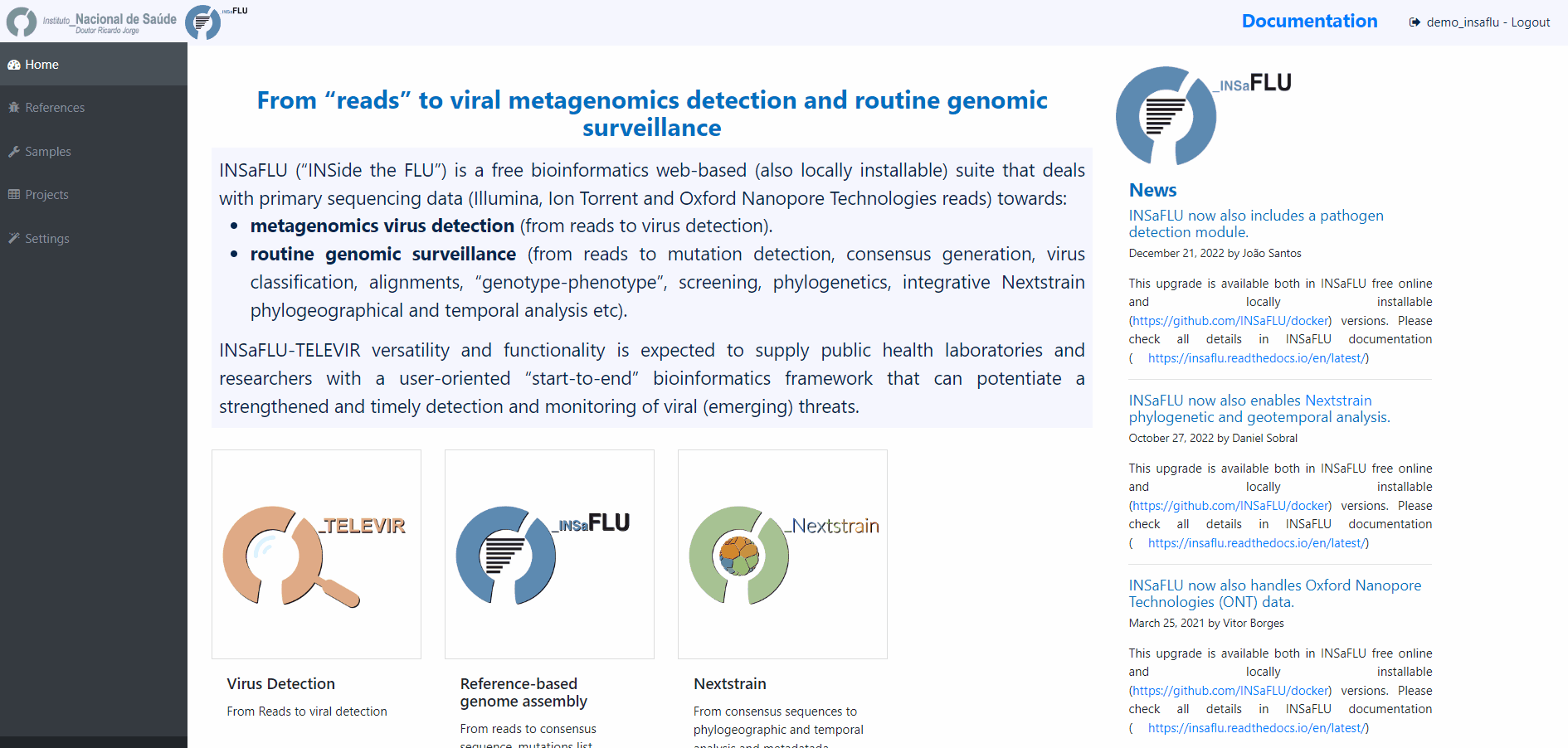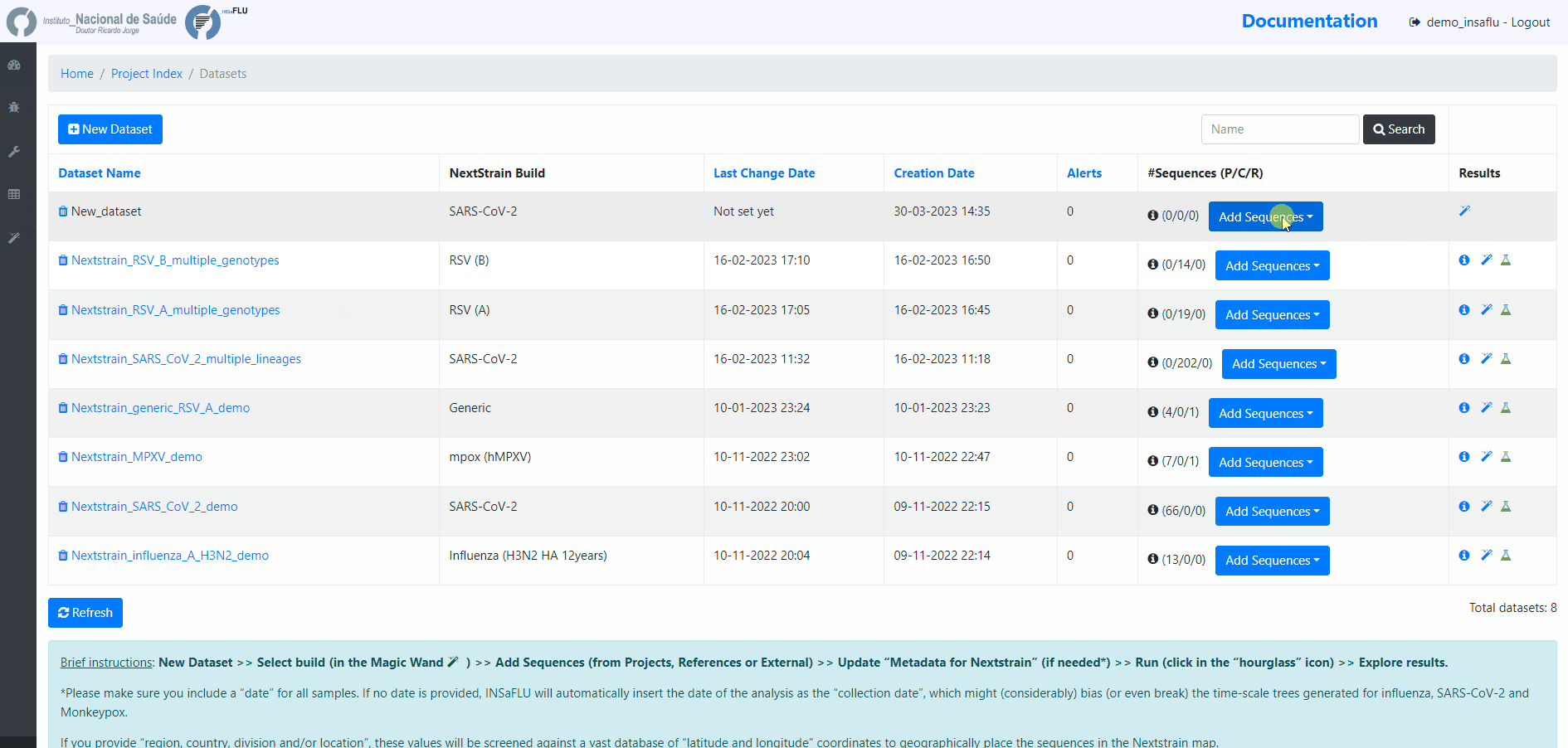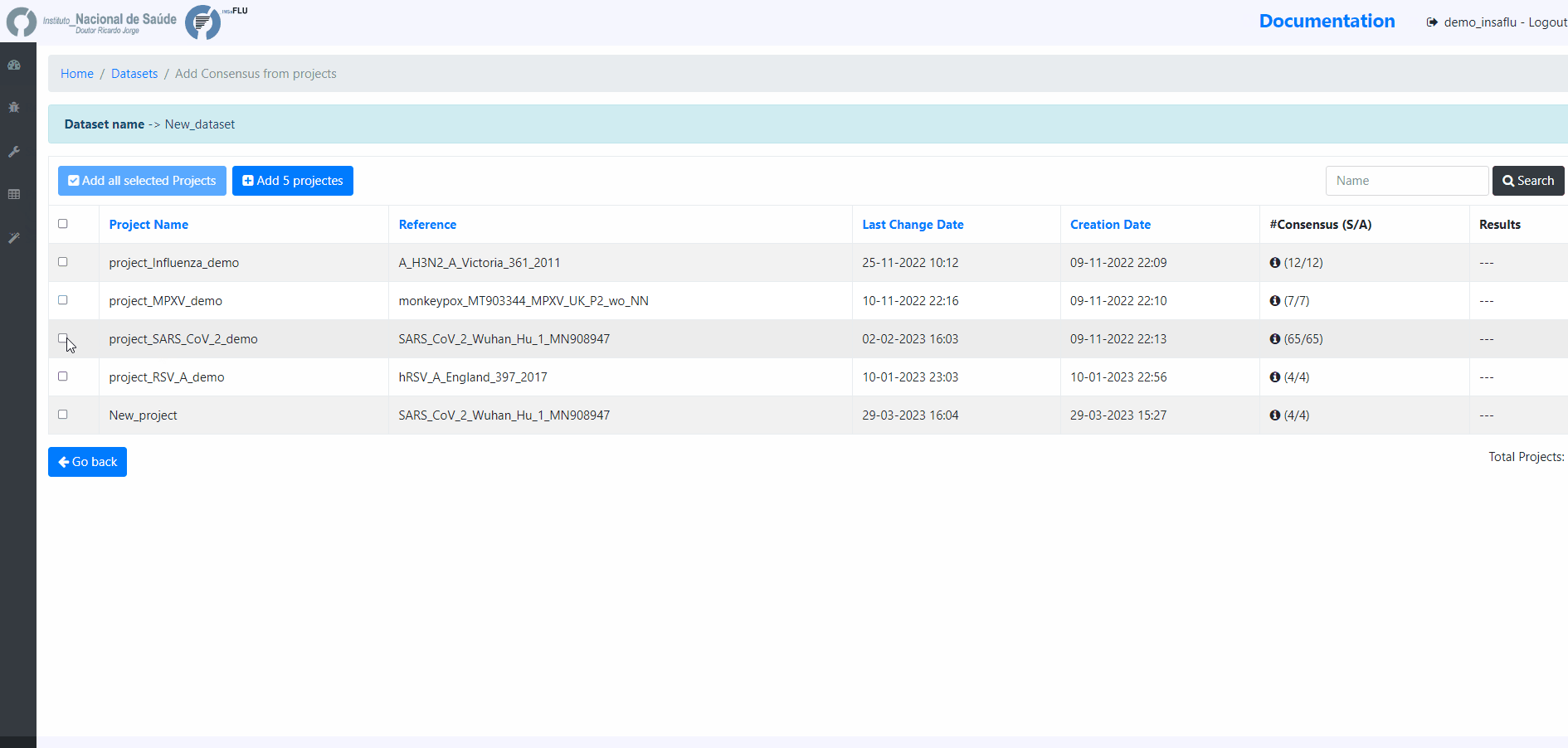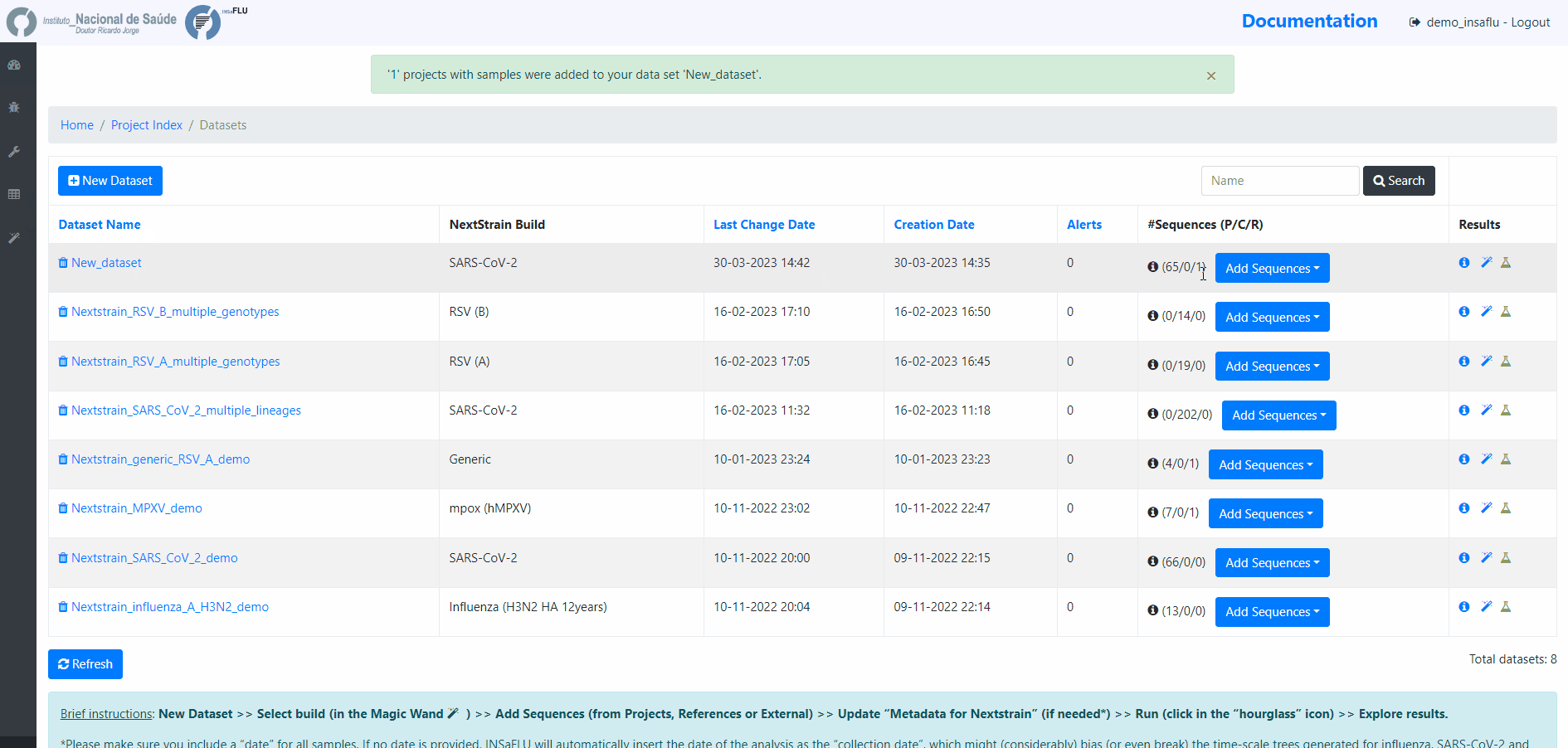
References - user-selected references sequences available in the References repository
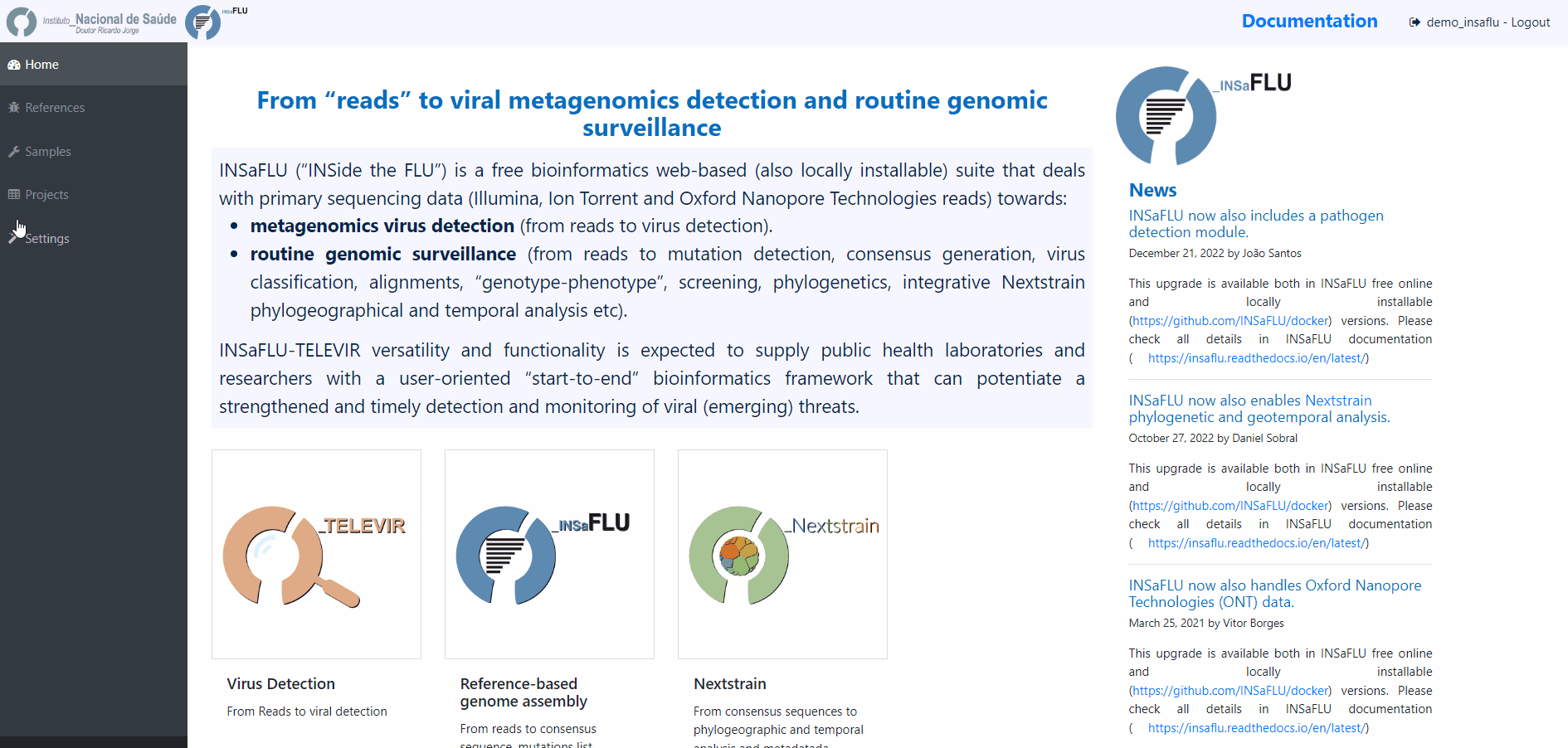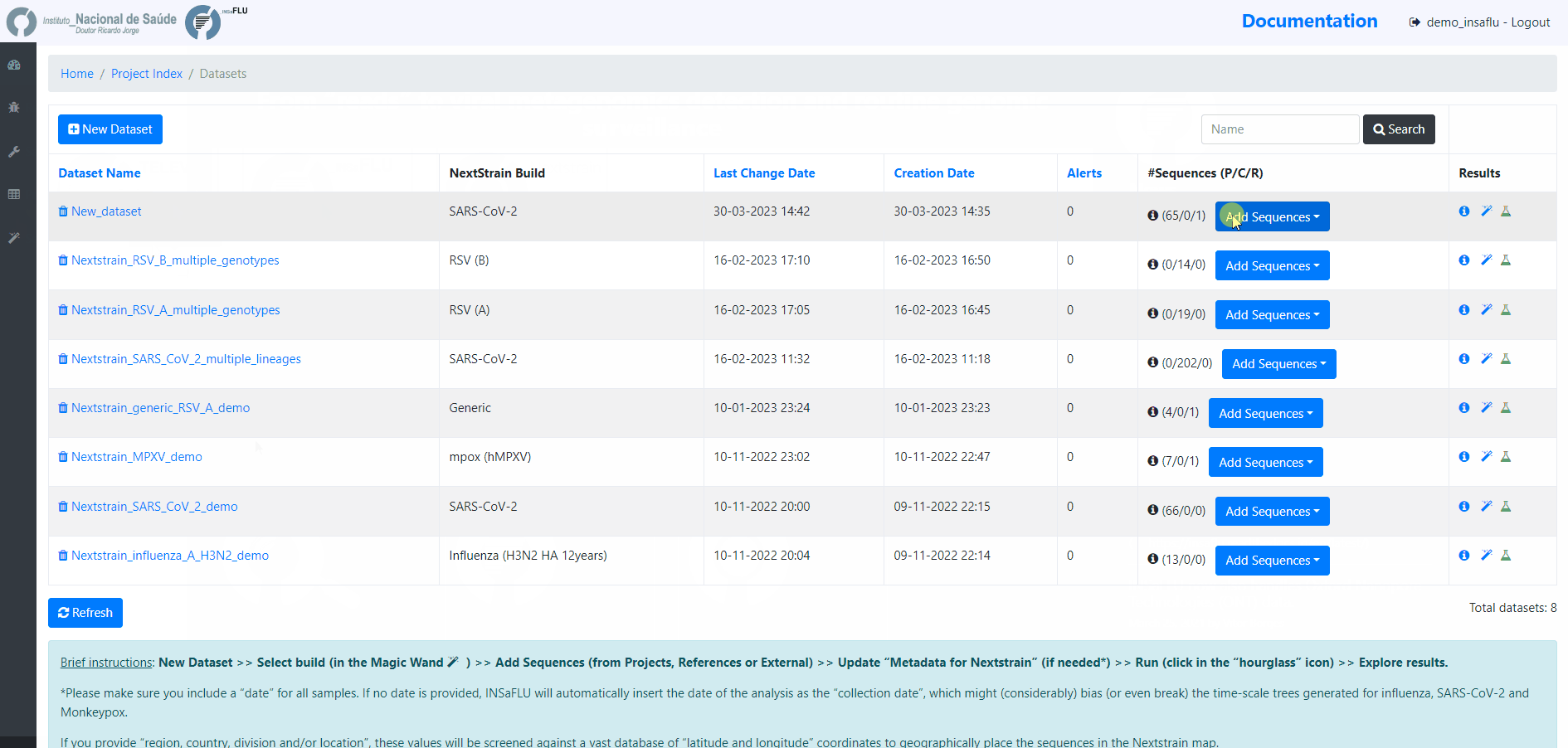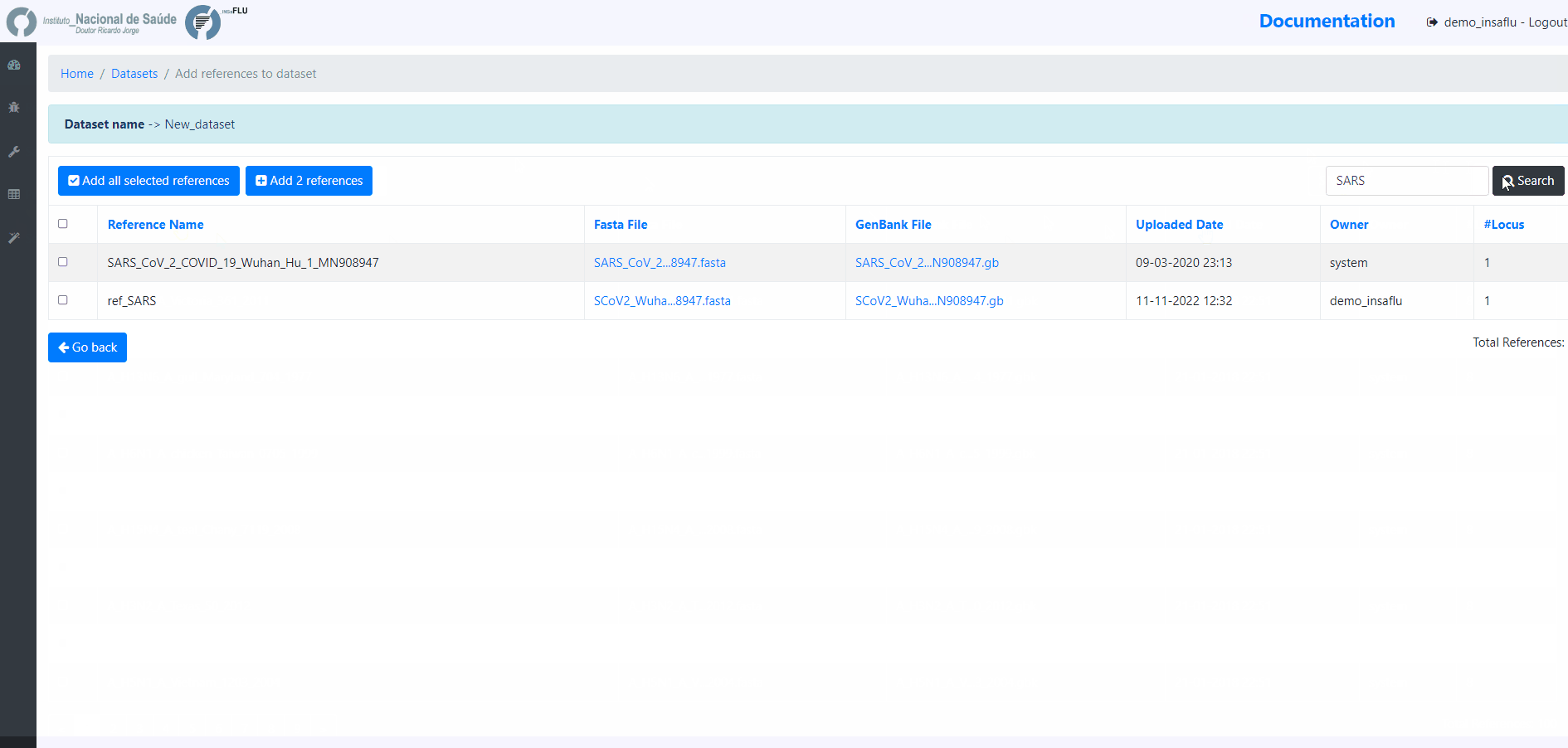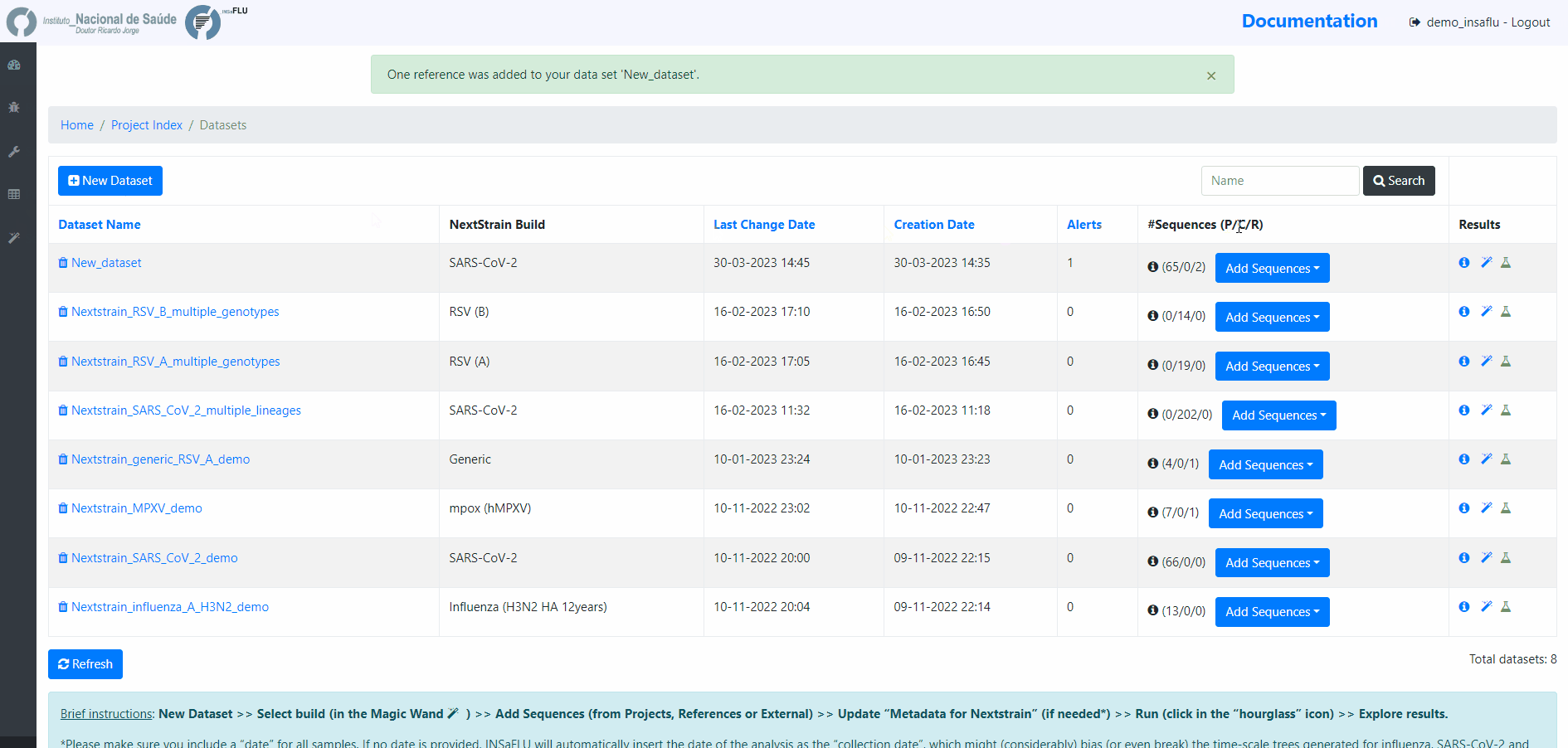
External sequences - to upload external sequences, click in “Add your onw consensus”, followed by “Upload new consensus”. You can upload FASTA or MULTI-FASTA files. Please make sure that the upload sequences match the respective build (e.g., genome sequences for SARS-CoV-2 Nextstrain build or HA sequences for influenza Nextstrain builds).
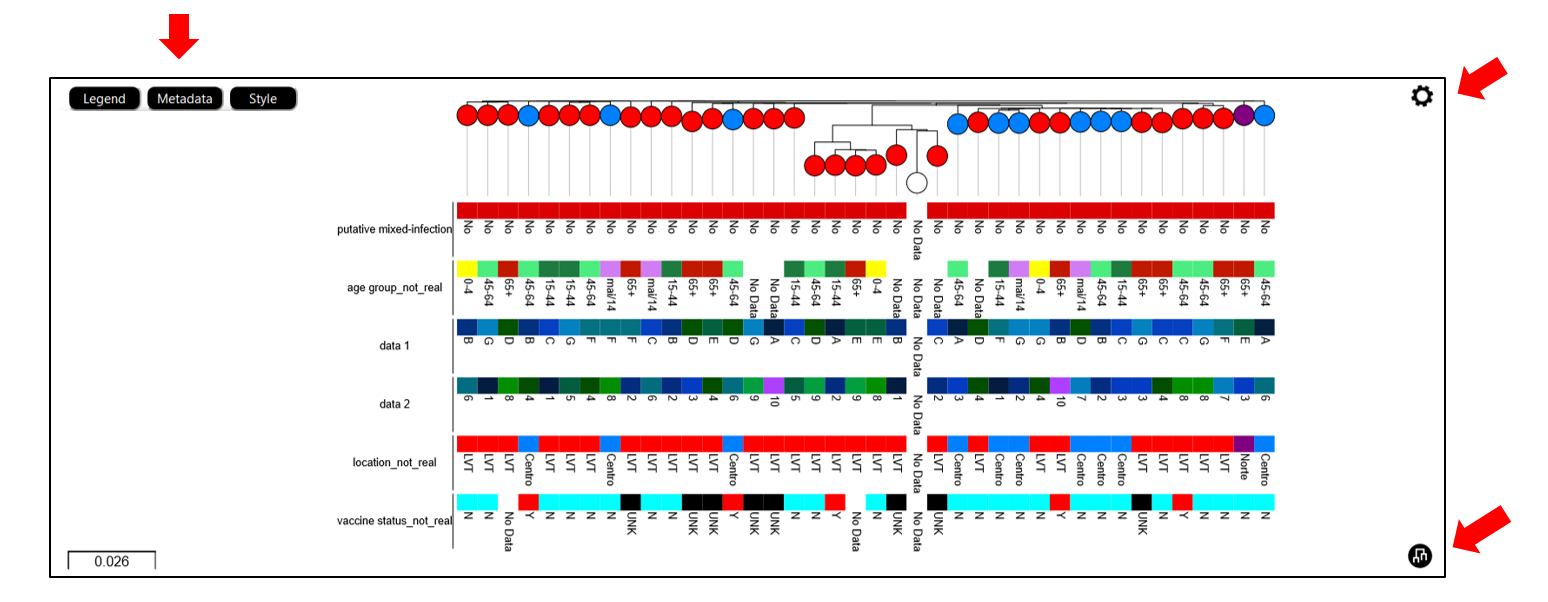
5. Enrich the metadata of the Dataset
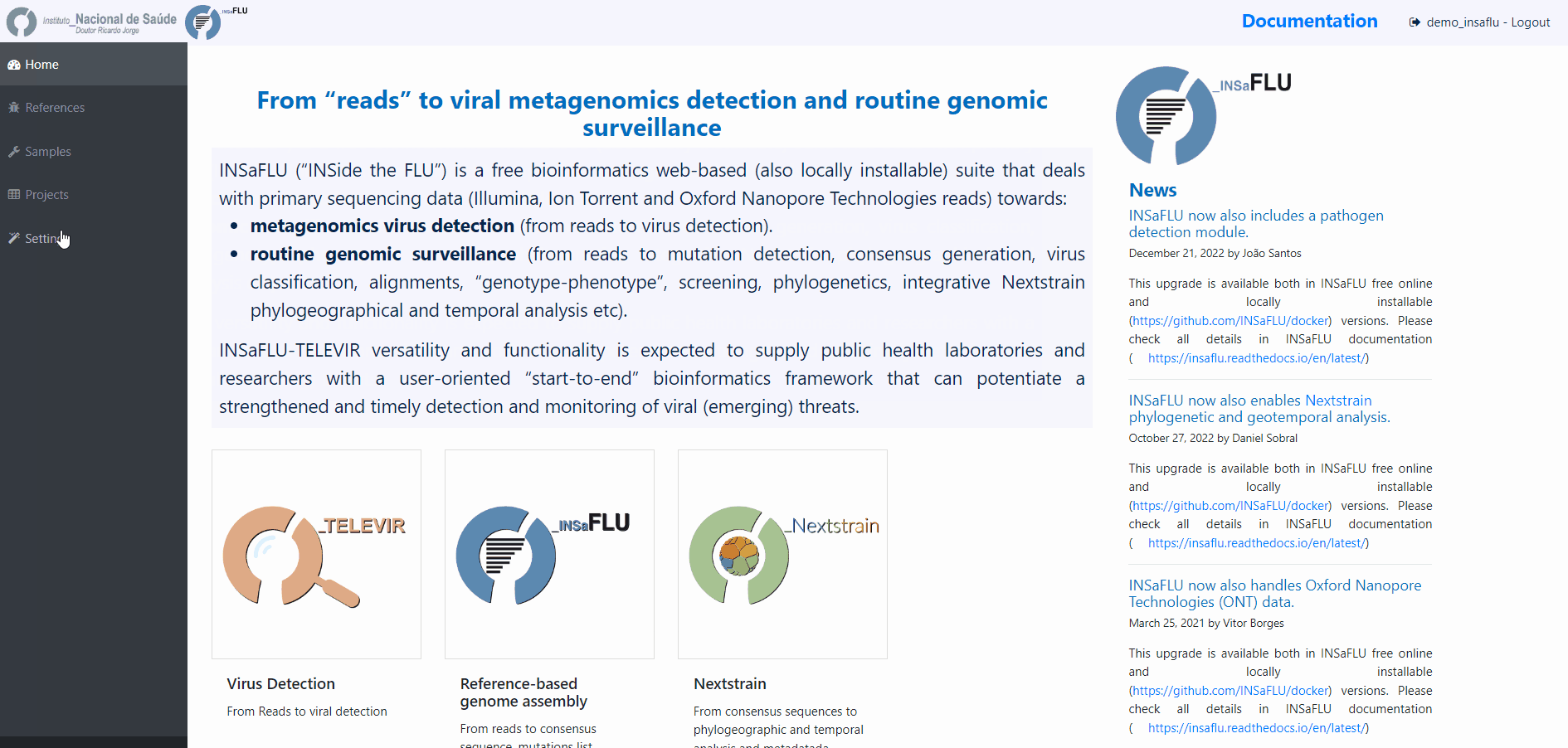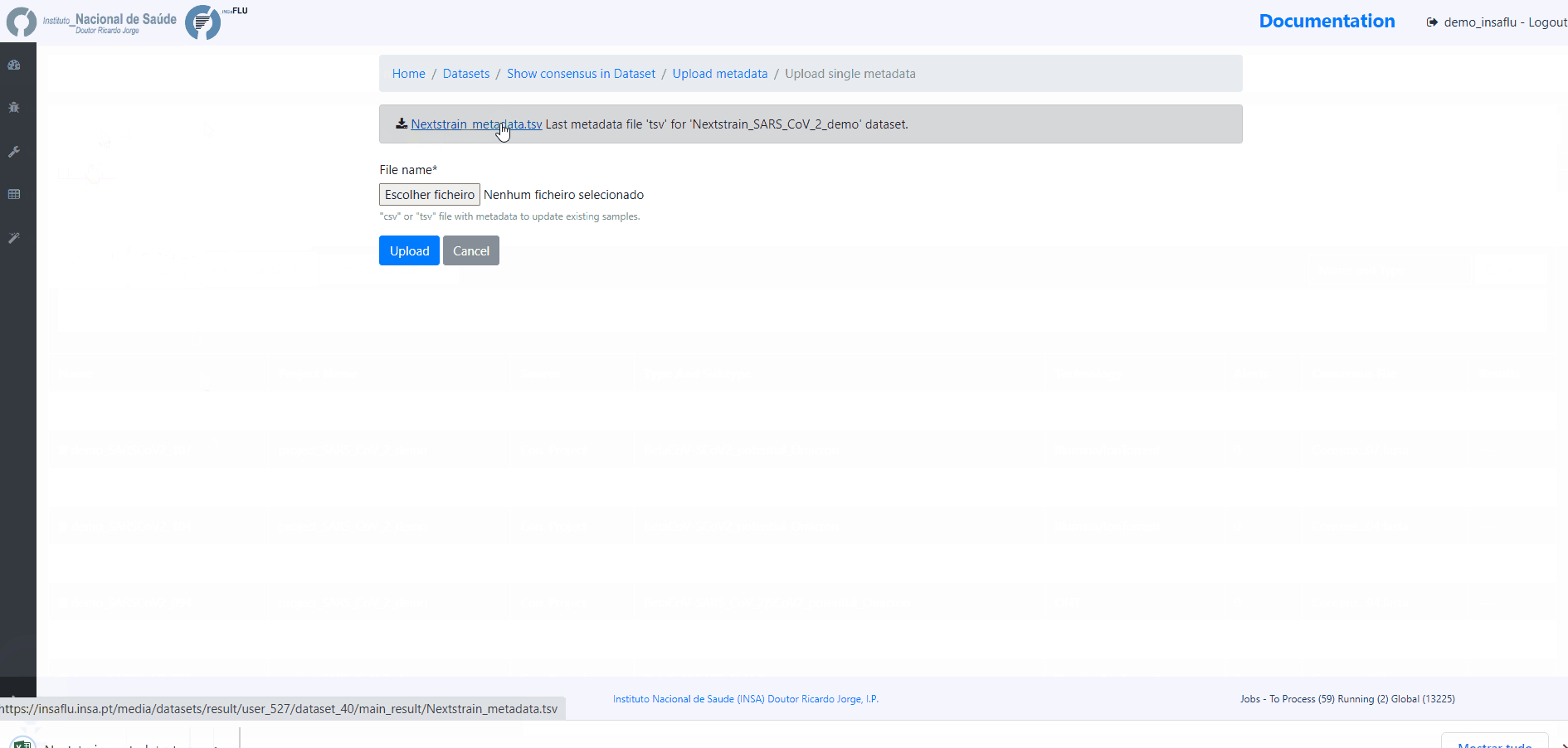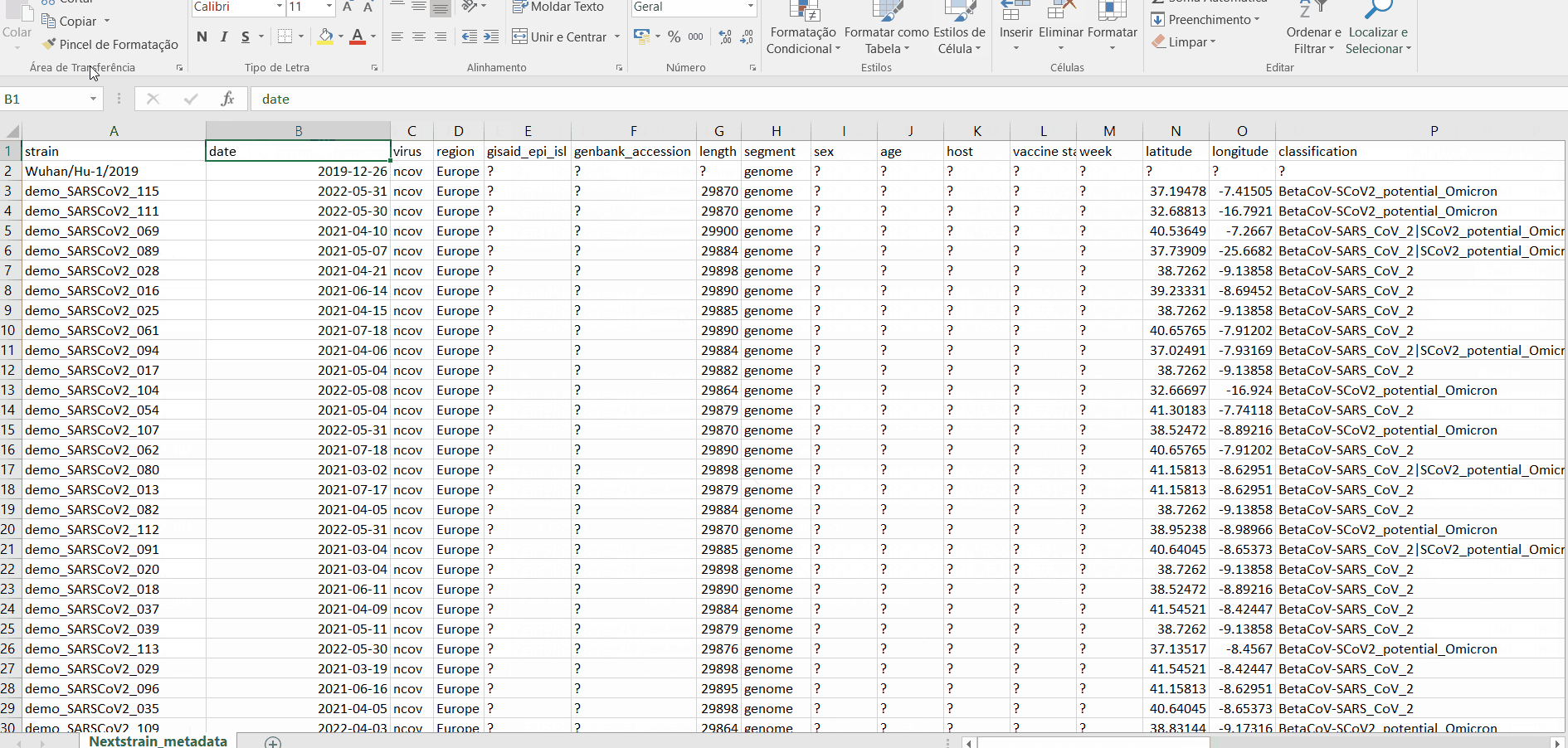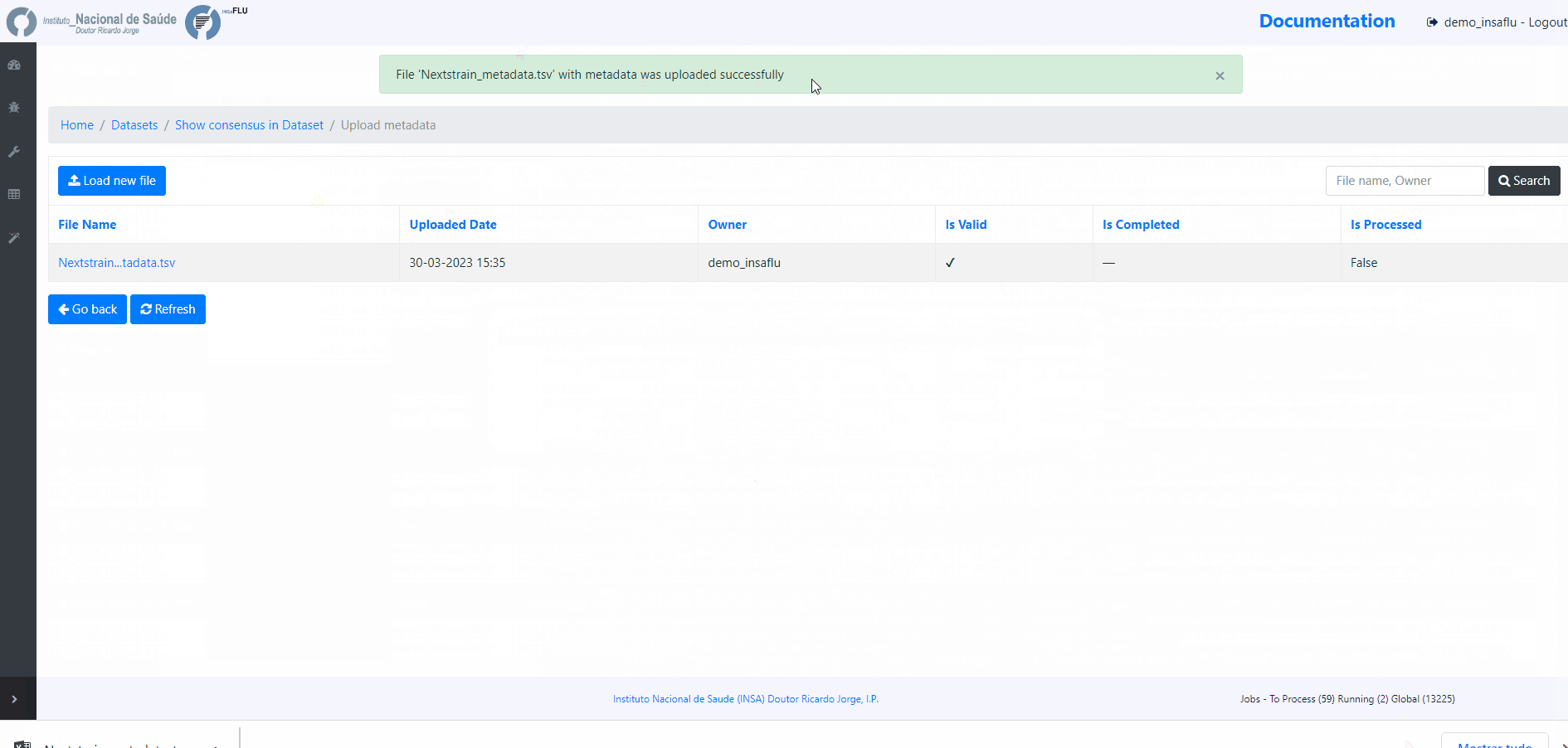
INSaFLU automatically prepares a “Nextstrain_metadata.tsv” table compiling all the metadata available for samples added to the Dataset. You can download the table, add more data and upload it again.
Important
To take advantage of temporal and geographical features of Nextstrain, please make sure you provide:
“collection date” for all samples added to Nextstrain datasets. If no collection date is provided, INSaFLU will automatically insert the date of the analysis as the “collection date”, which might (considerably) bias (or even break) the time-scale trees generated for influenza, SARS-CoV-2 and Monkeypox.
“latitude” and “longitude” AND/OR “region”, “country”, “division” and/or “location” columns in the metadata. These values will be screened against a vast database of “latitude and longitude” coordinates (https://github.com/INSaFLU/nextstrain_builds/blob/main/generic/config/lat_longs.tsv) to geographically place the sequences in the Nextstrain map.
To update the Nextstrain metadata of a given Dataset, please click in “Metadata for Nextstrain”, download the previous table, update it with new data and upload it. Then, click in the “hourglass” icon to Rebuild the Nexstrain outputs.
Note: For sequences previously obtained with INSaFLU (i.e., consensus sequences inported to “Datasets” from the “Projects” module), you can also add/update the metadata following these instructions: https://insaflu.readthedocs.io/en/latest/uploading_data.html#updating-sample-metadata (this option is not available for external sequences).
6. Run your Dataset
After adding samples, click in the “hourglass” icon to start the Nexstrain analysis.
5. Scale-up your Dataset.
You may add more samples to your Dataset at any time and click in the “hourglass” icon to Rebuild the Nexstrain outputs.
6. Remove samples from your Datasets.
You may remove some samples from your Dataset.
Output Visualization and Download
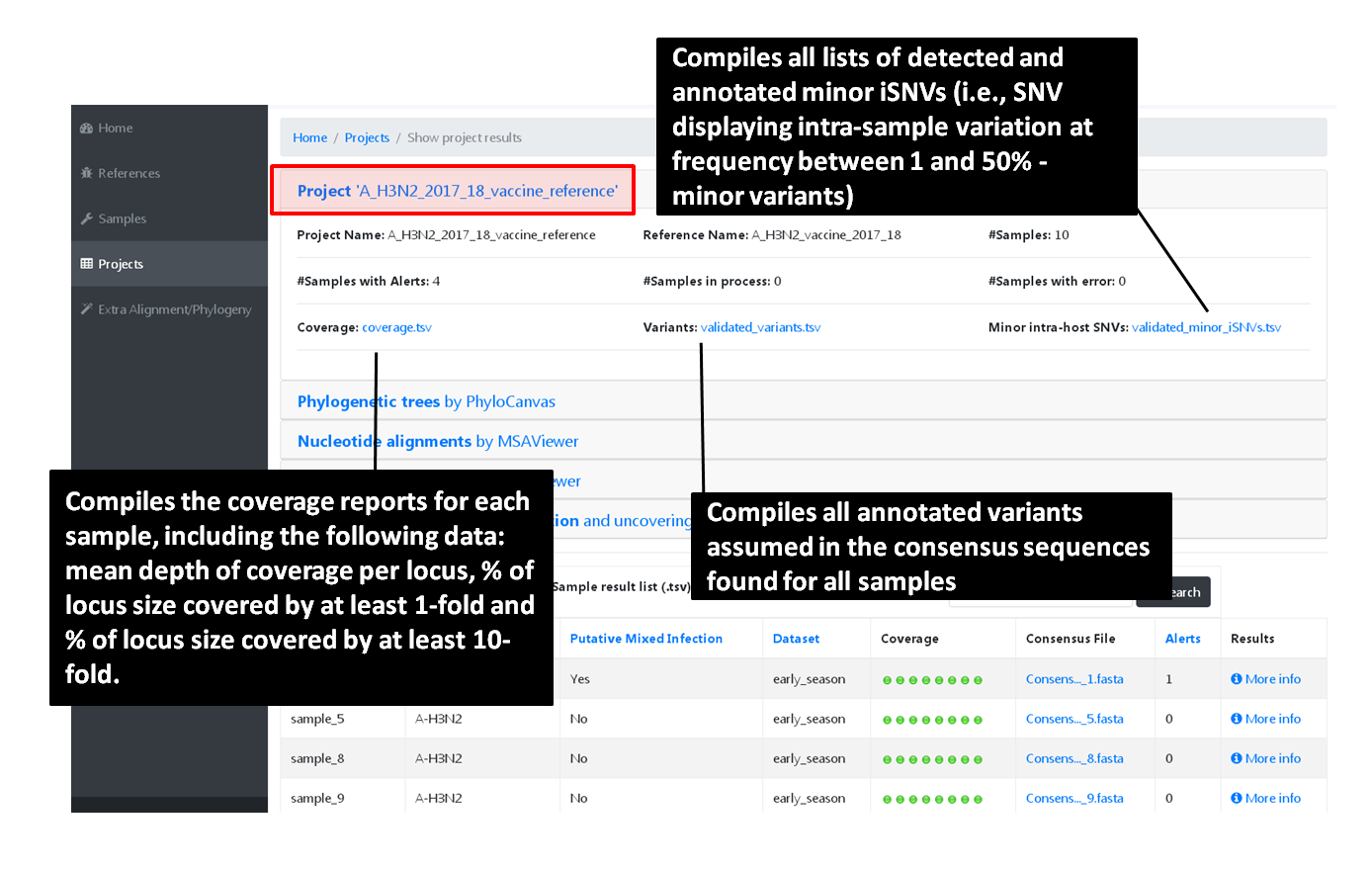
The surveillance-oriented INSaFLU component generates multiple outputs, which include:
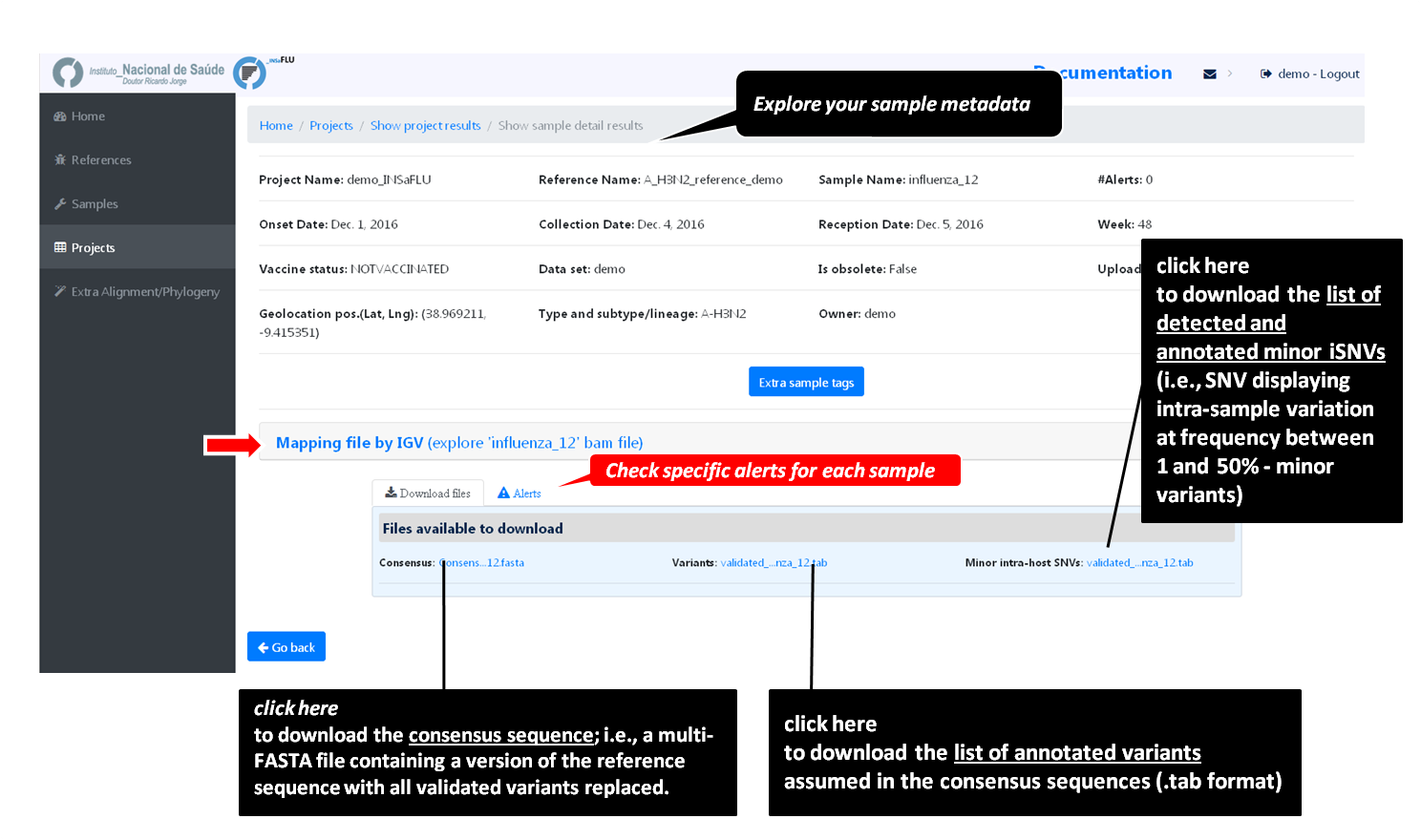
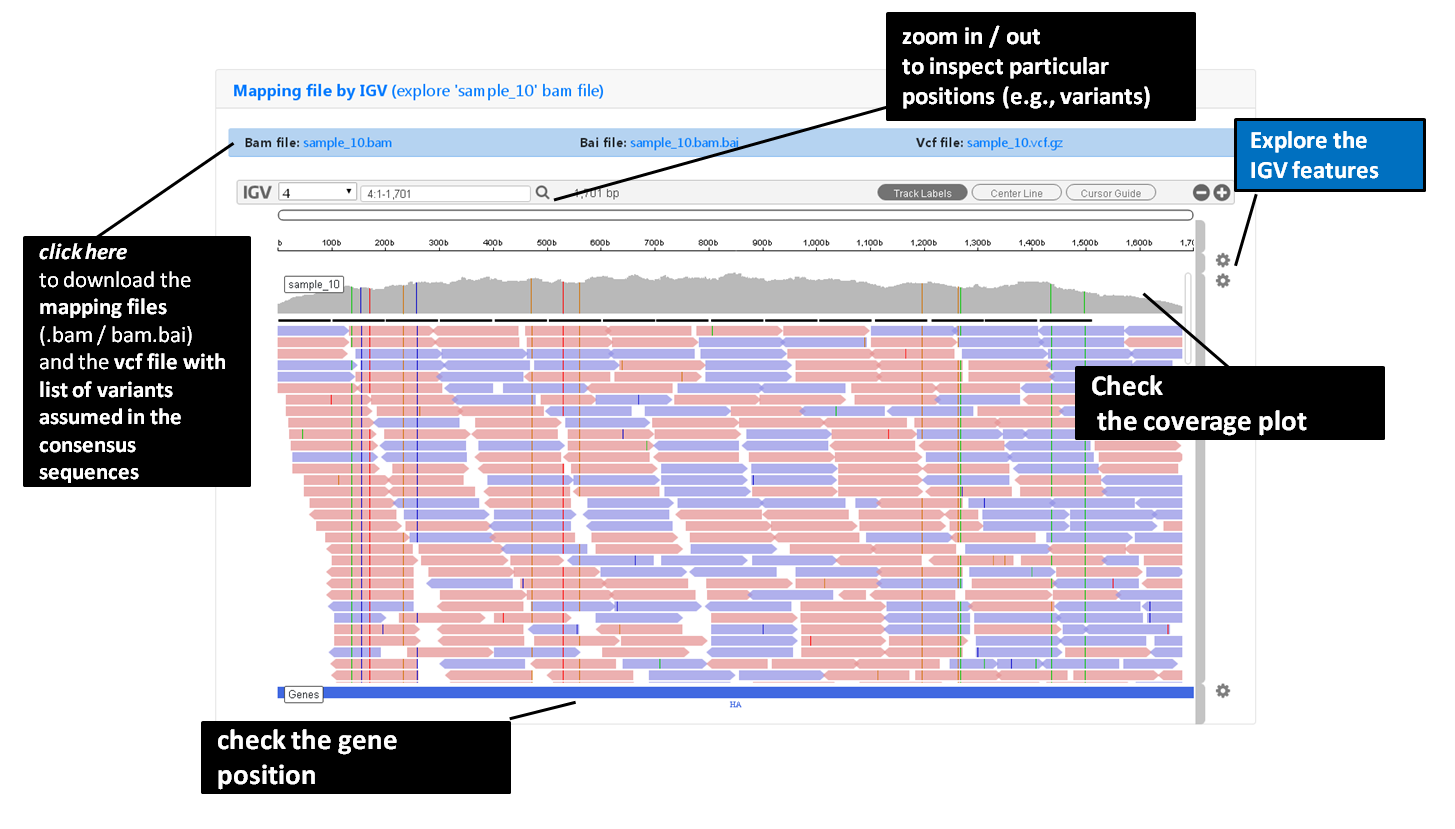
sample-specific outputs (such as, QC reports, mapping files, mutation annotation and consensus sequences)
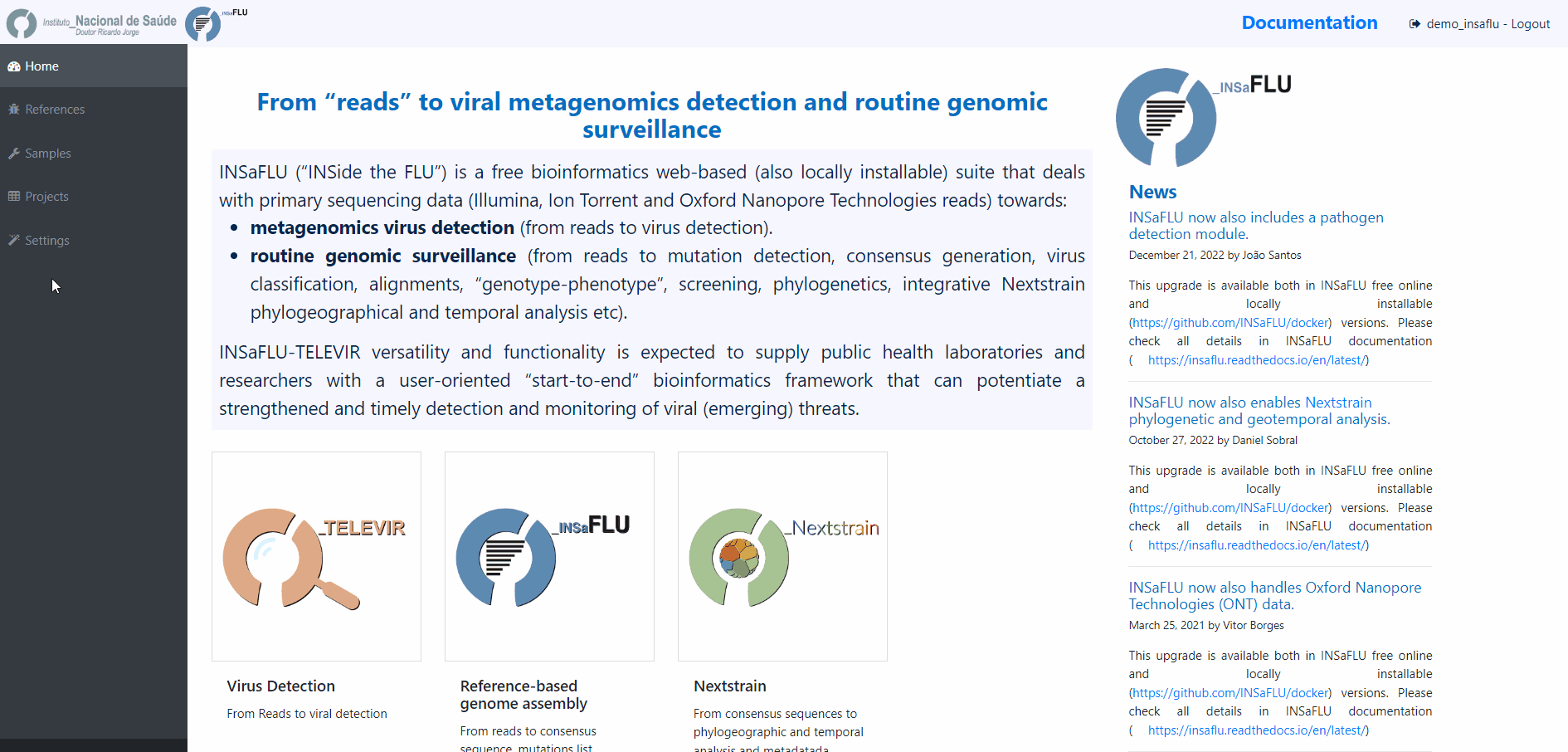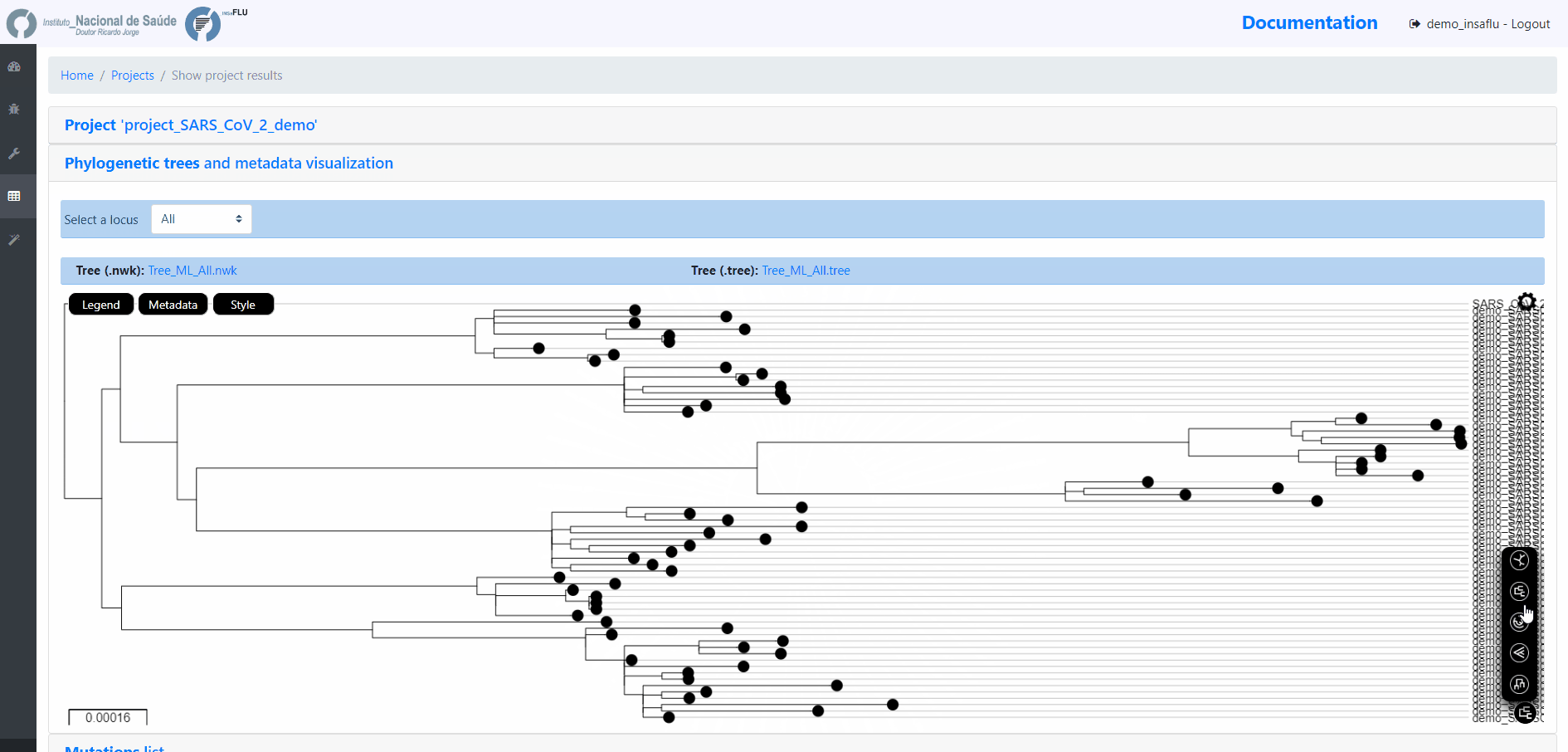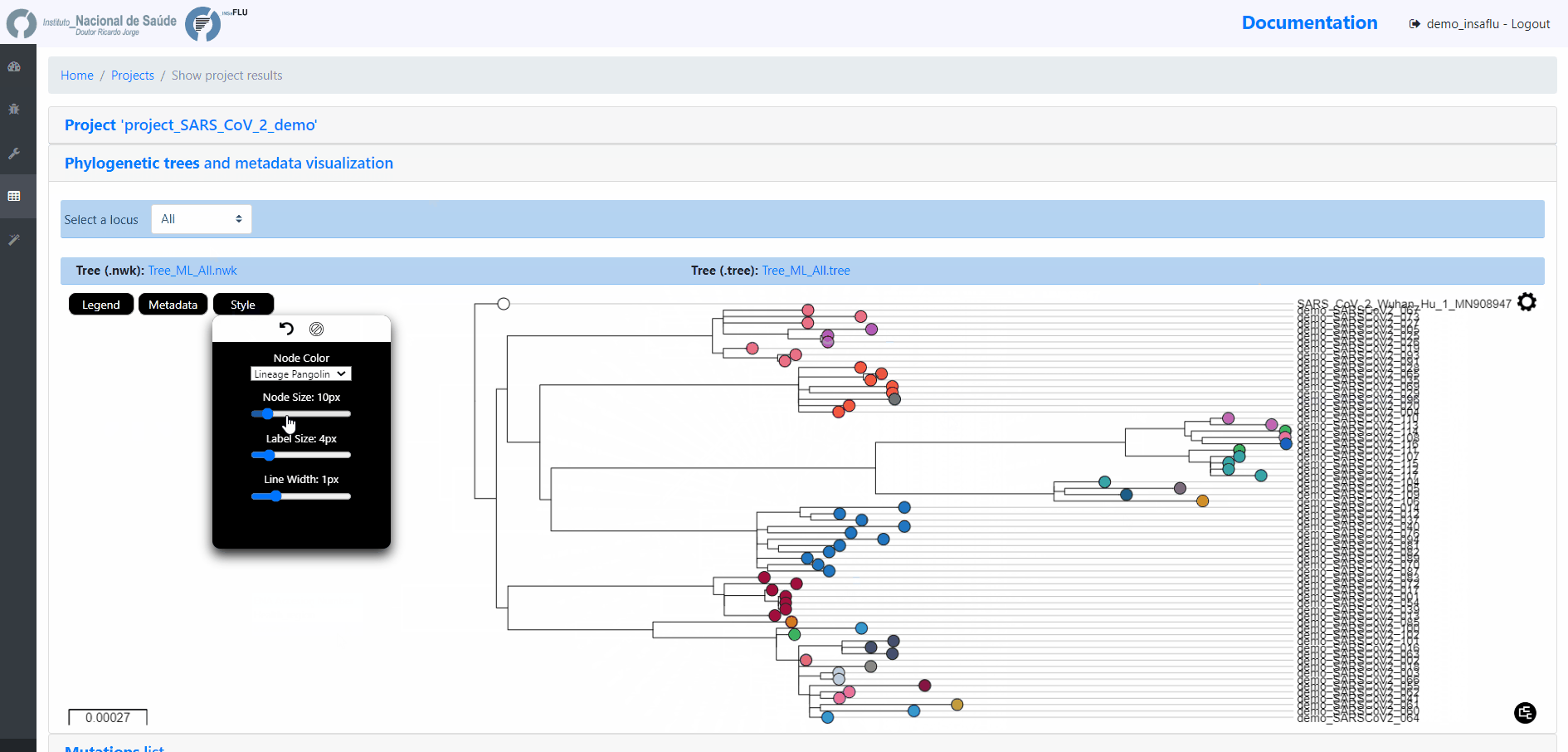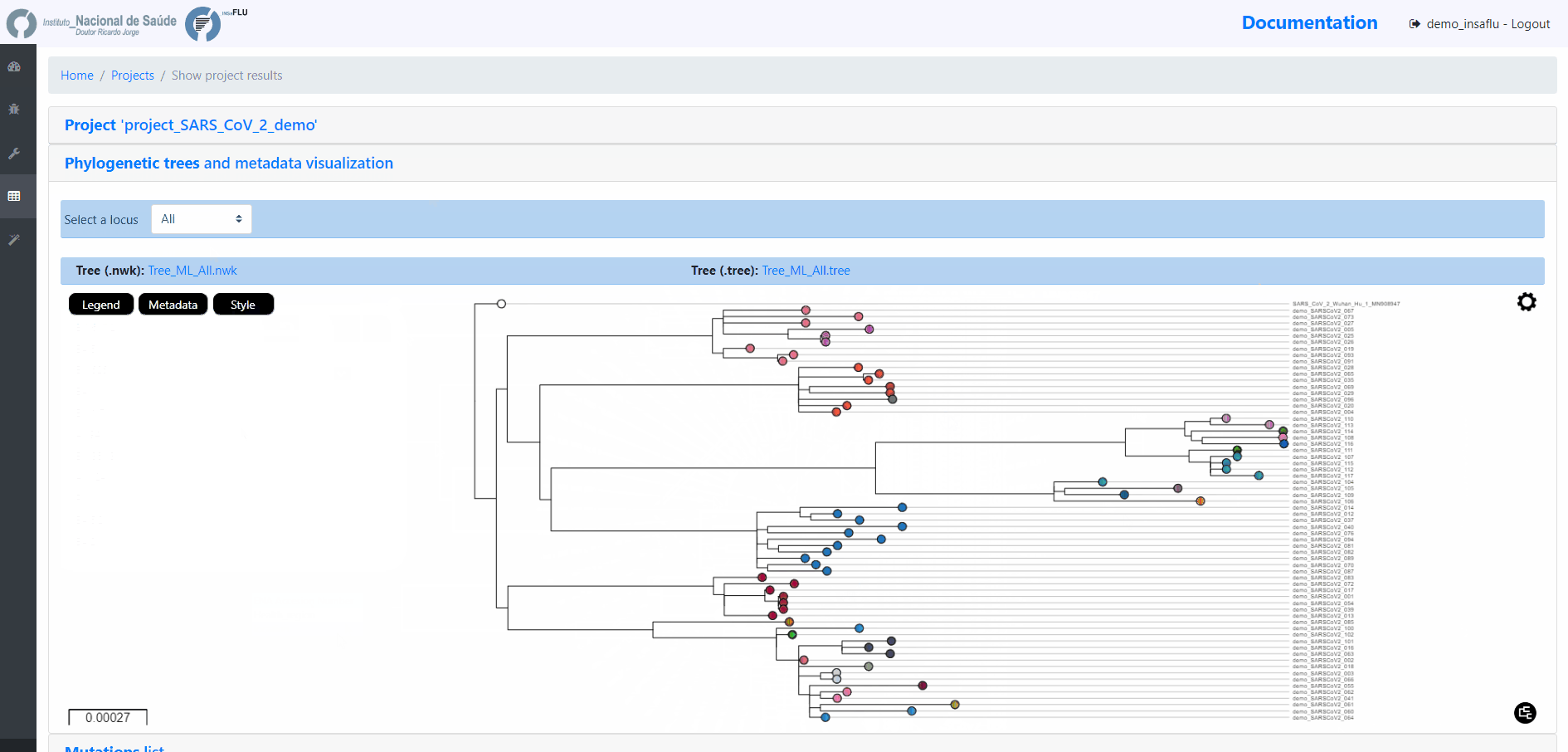
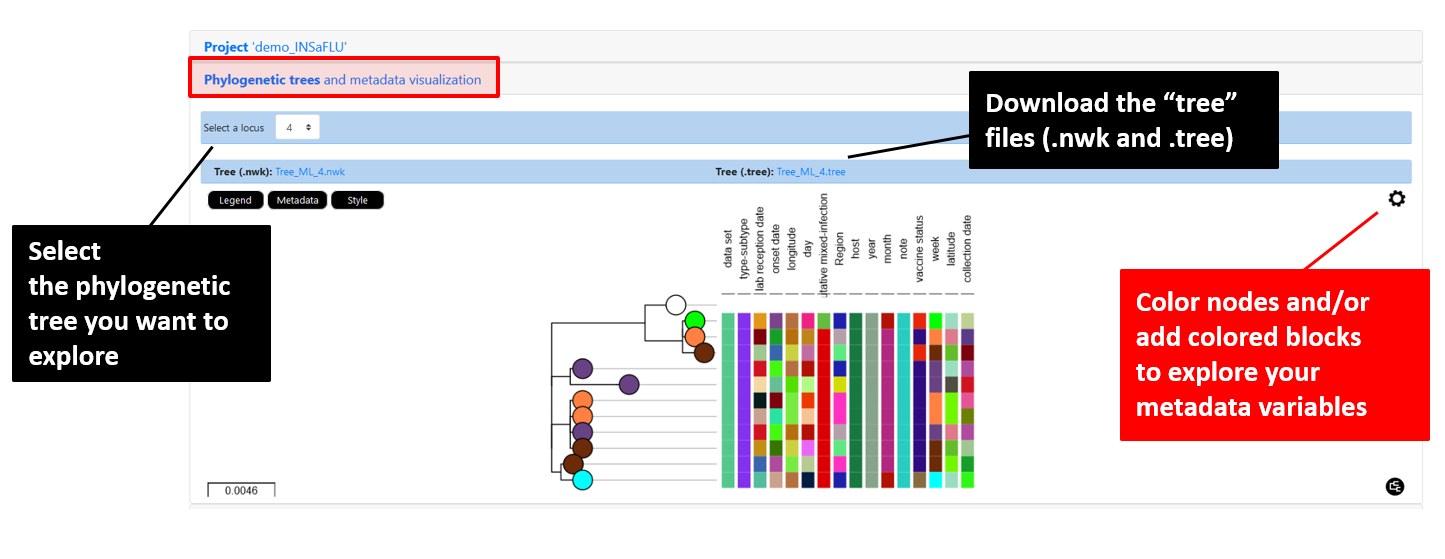
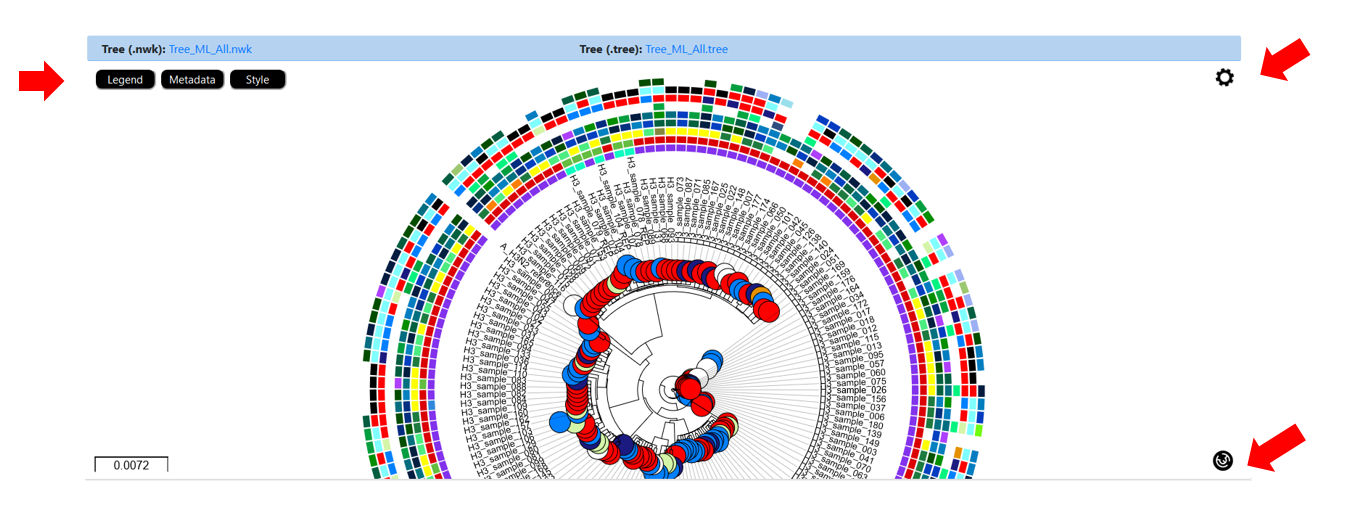
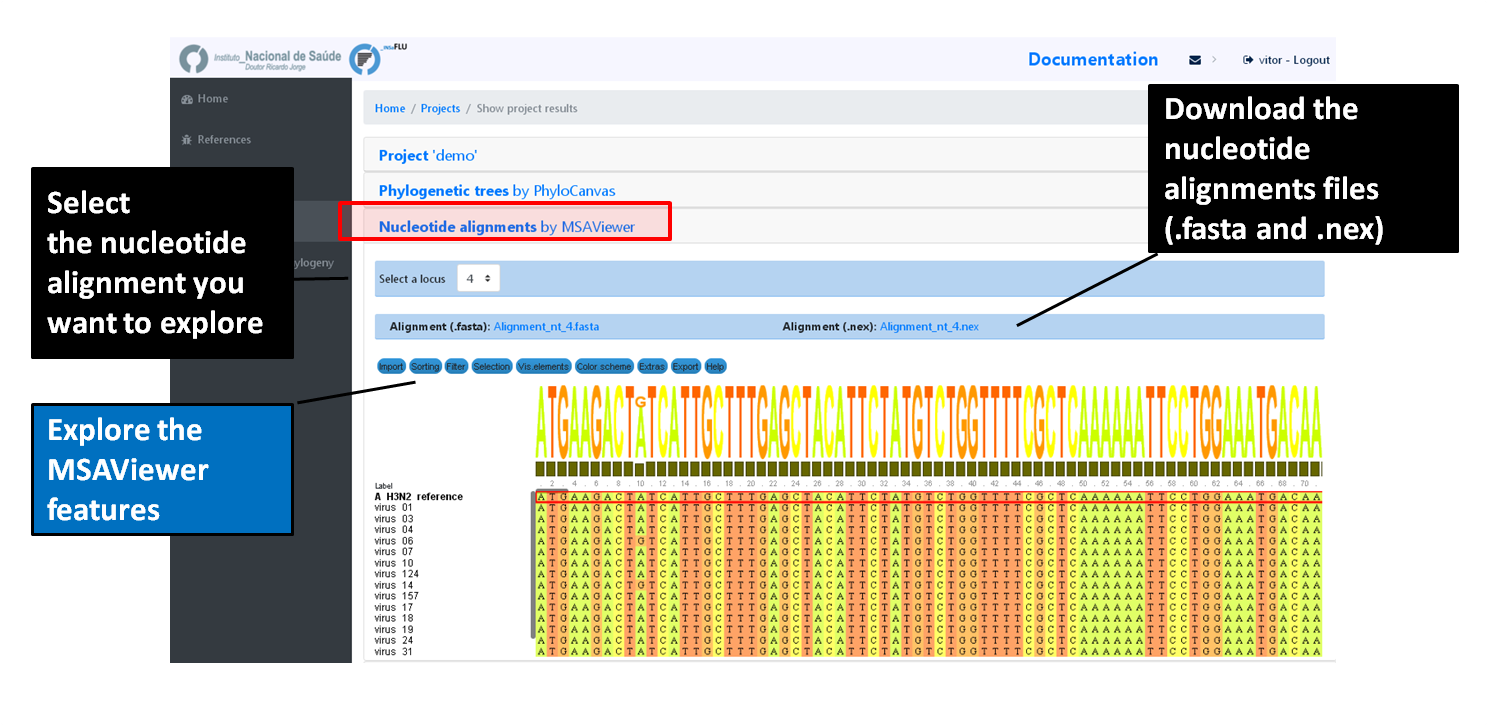
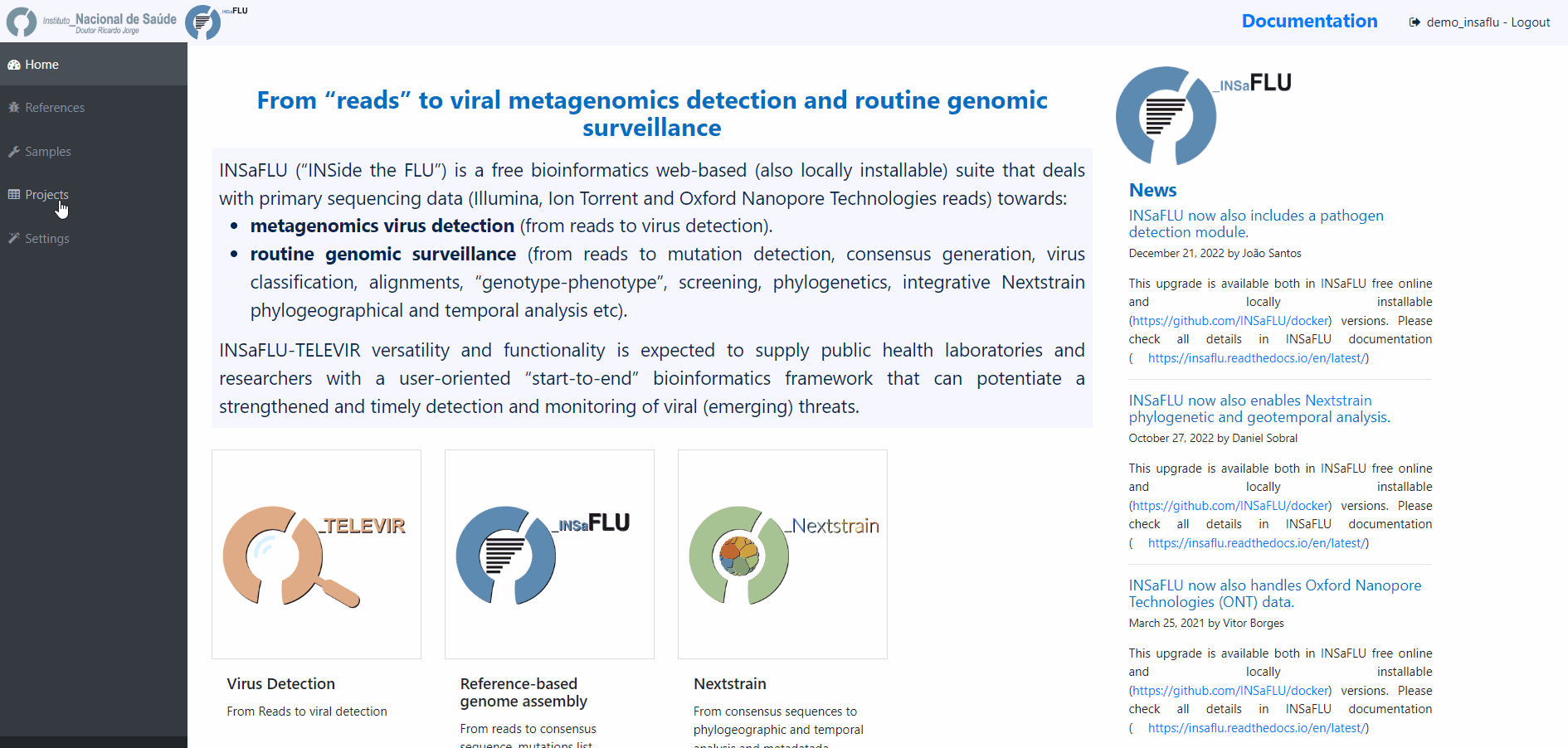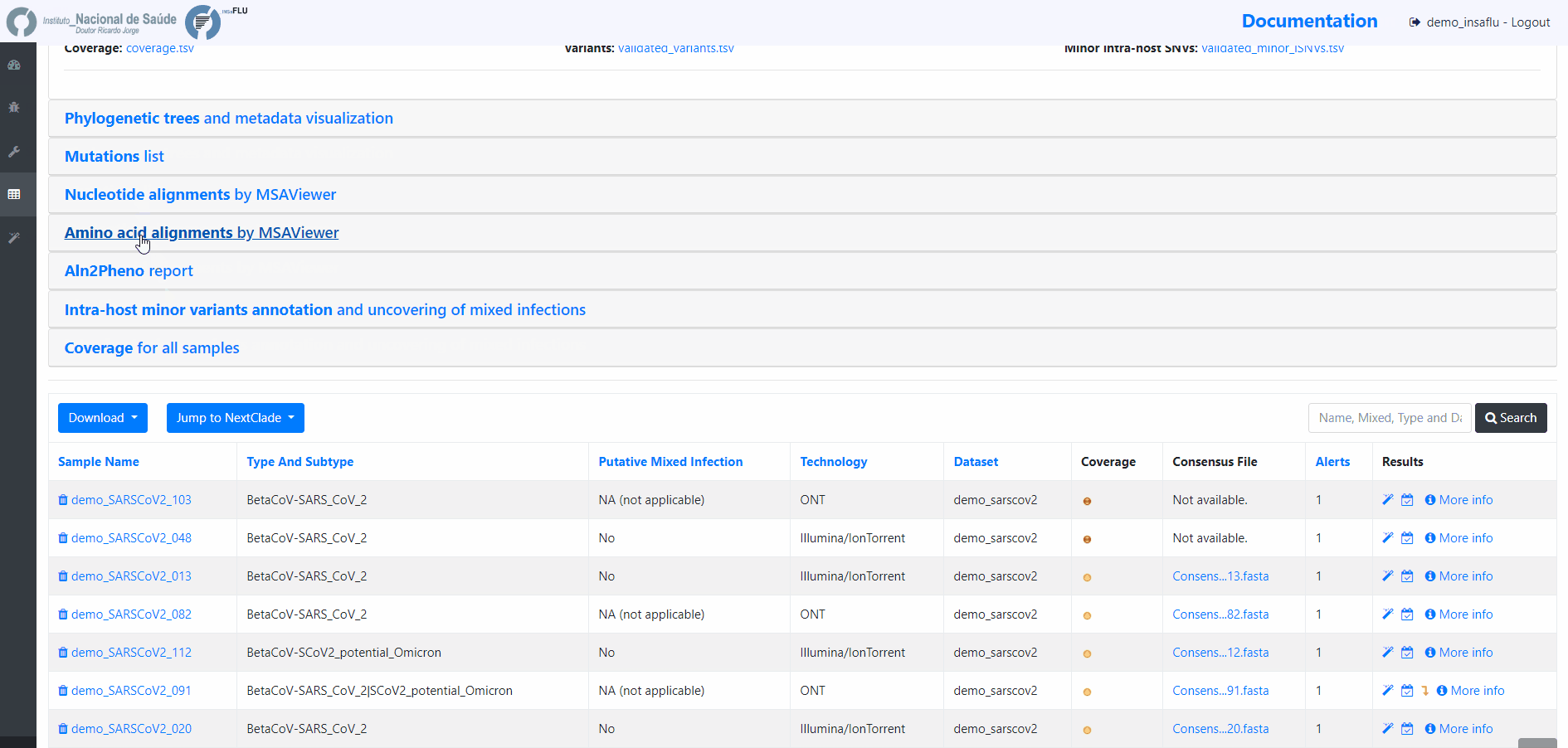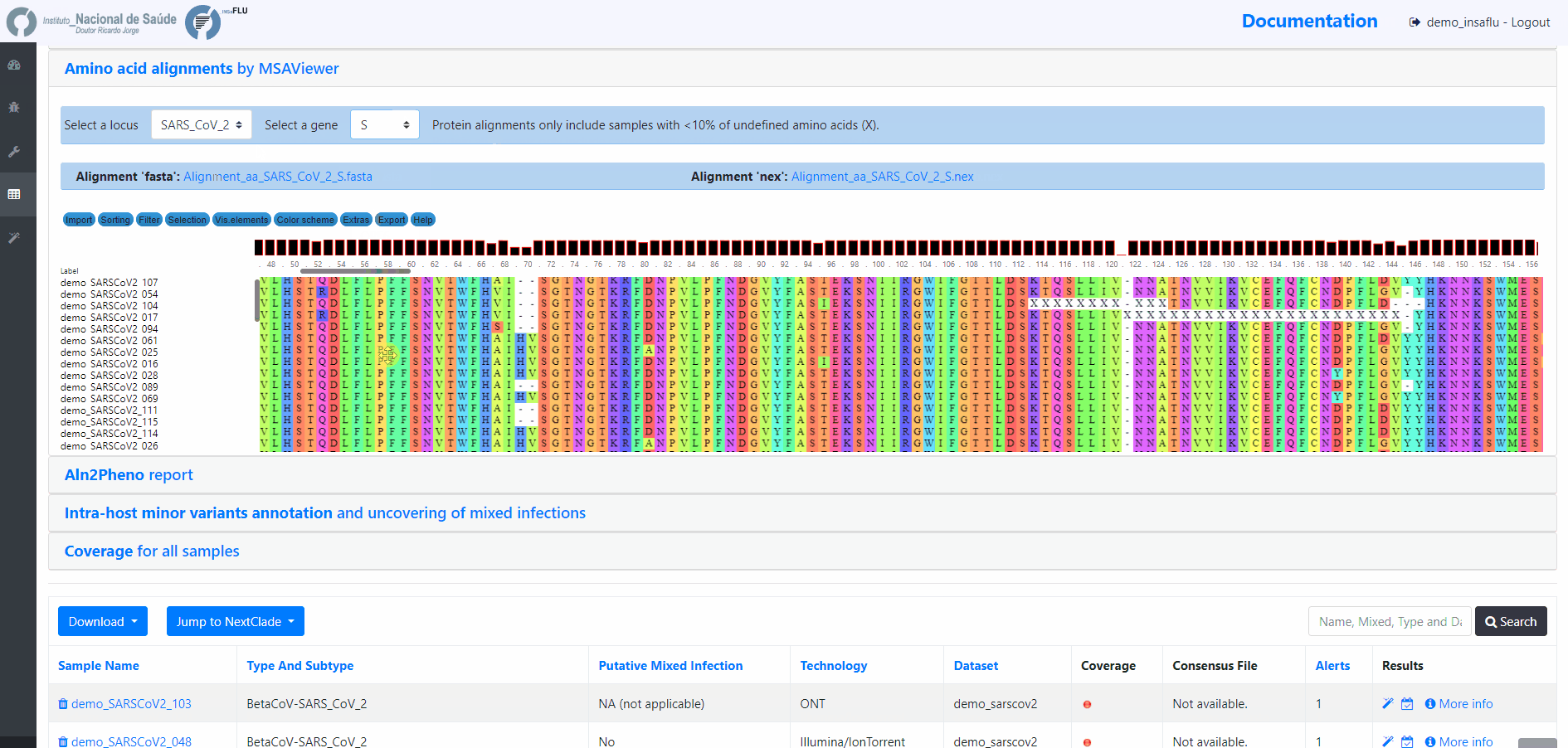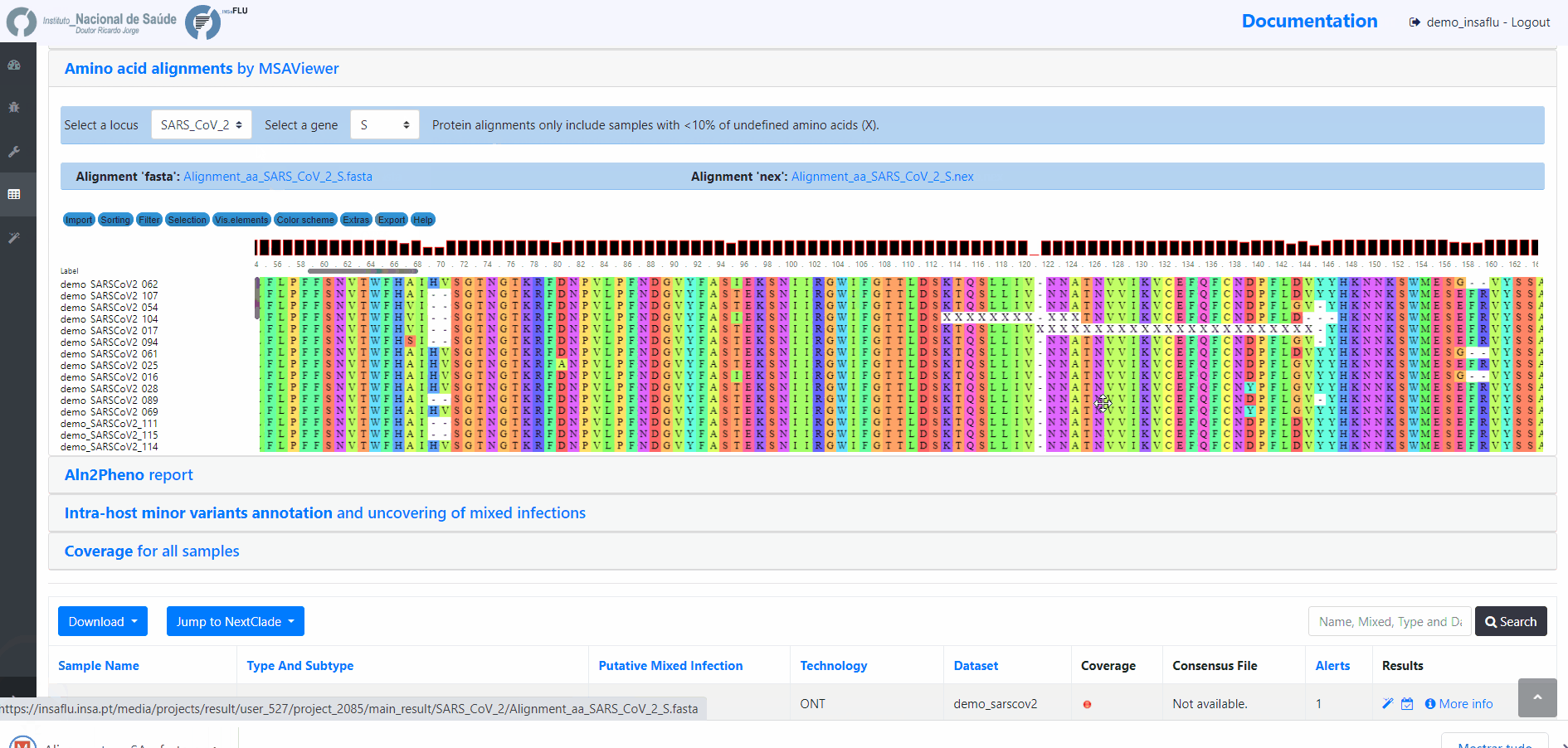
INSaFLU Project outputs (such as, nucleotide/amino acid alignments and phylogenetic trees).
Nextstrain Dataset outputs (such as, Nextstrain alignments and integrative phylogeographical and temporal data).
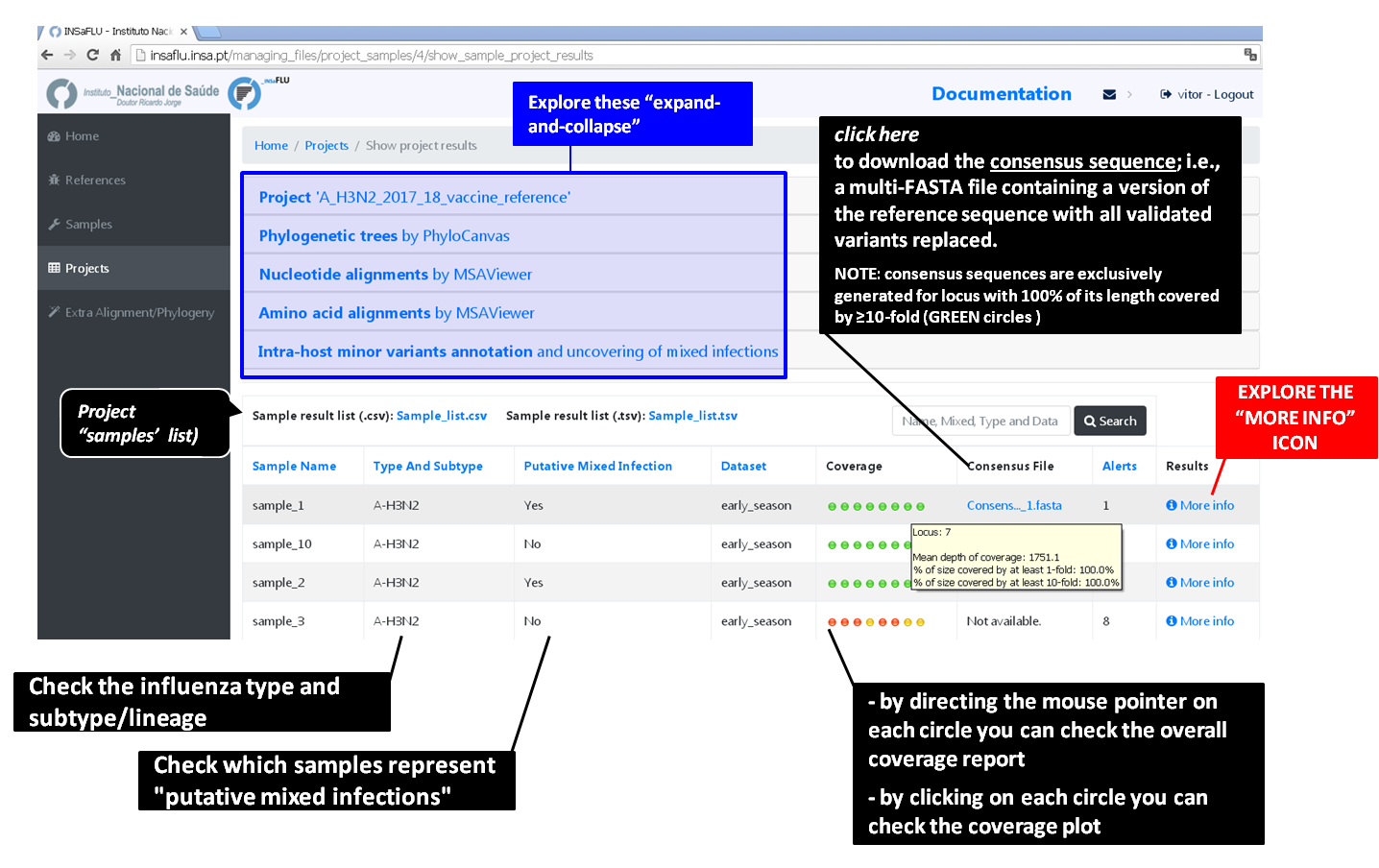
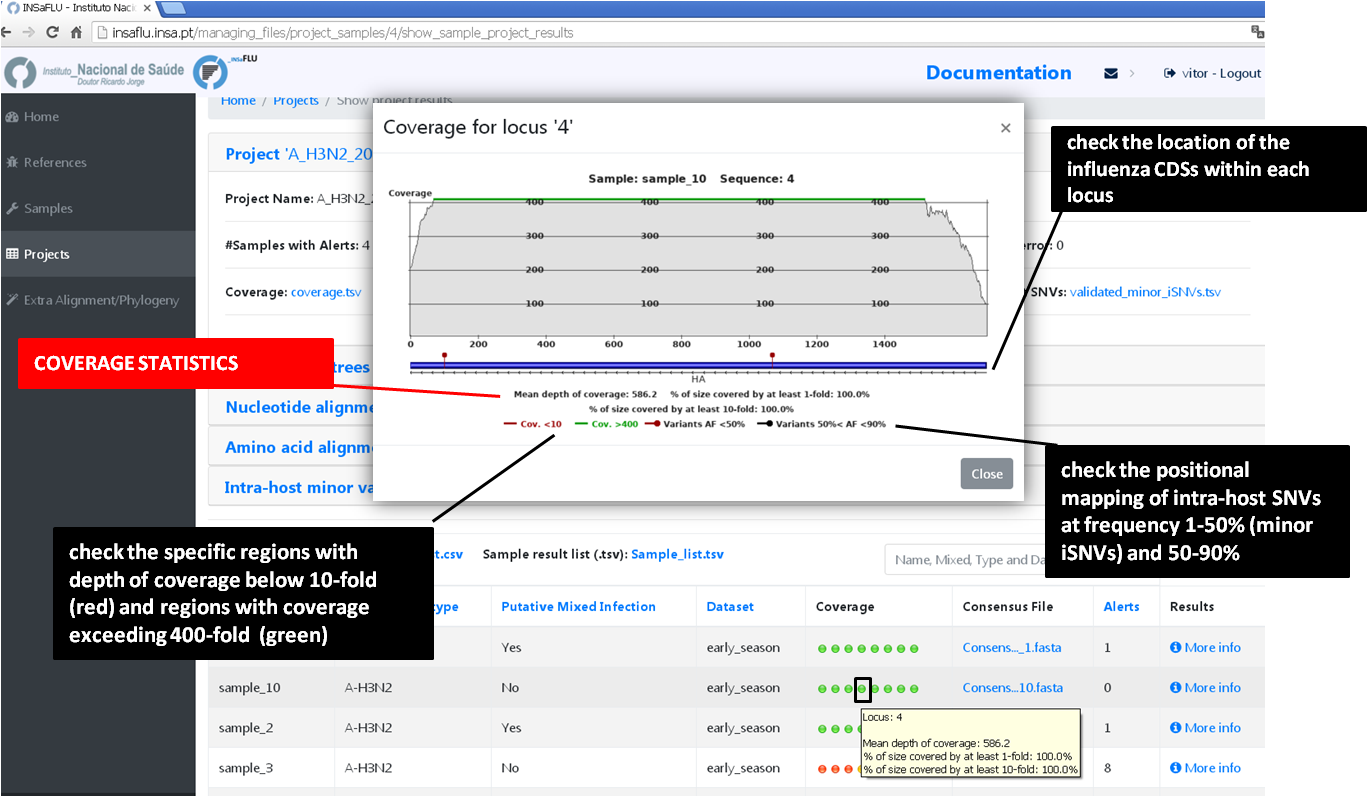
Outputs are organized by the dynamic “expand-and-collapse” panels that allow you a user-friendly visualization/download of all graphical, text and sequence output data. The following table provides an overview on all INSaFLU outputs organized by bioinformatics module:
INSaFLU_current_outputs_08_09_2023.xlsx
While navigating through INSaFLU menus, you will find which main software (including versions and settings) were used to generate outputs. The Sample list of each Project also also summarizes the software settings and user-defined cut-offs applied for each sample.