Uploading or Updating data
INSaFLU needs:
NGS data (fastq reads) (mandatory)
Sample metadata (to link each sample to the respective NGS data) (mandatory)
Reference data (additional user-restricted reference sequences) (optional)
Uploading Sample metadata and NGS data
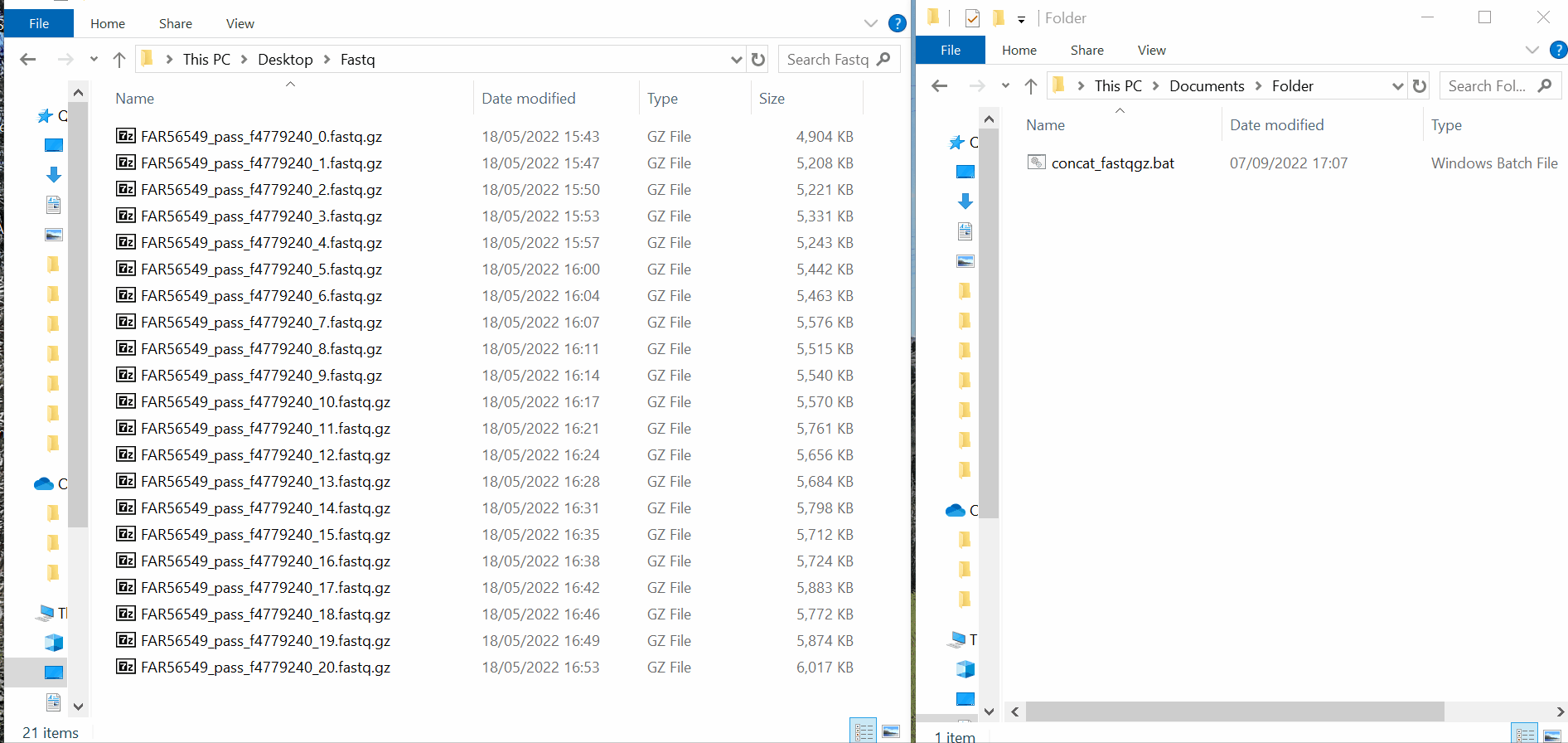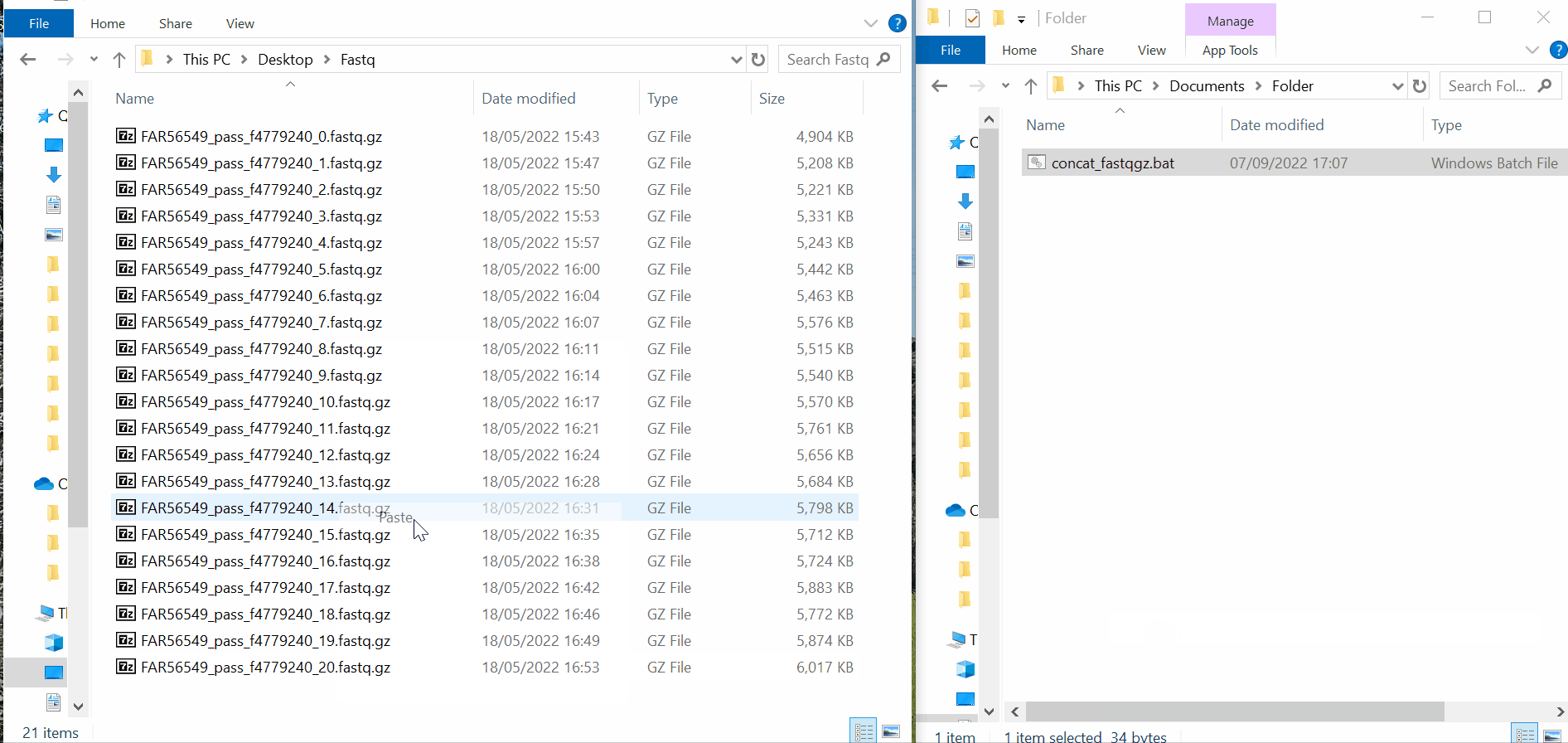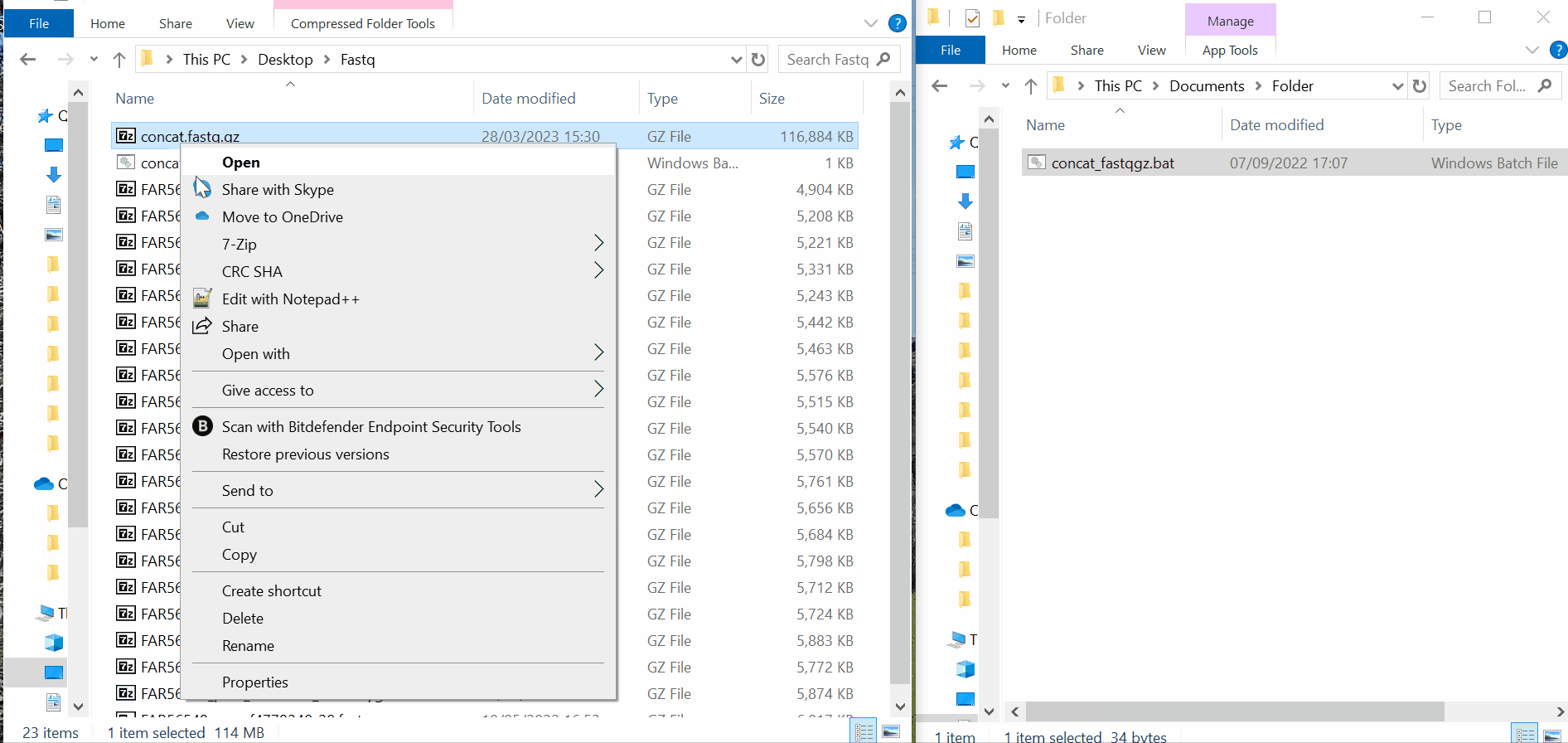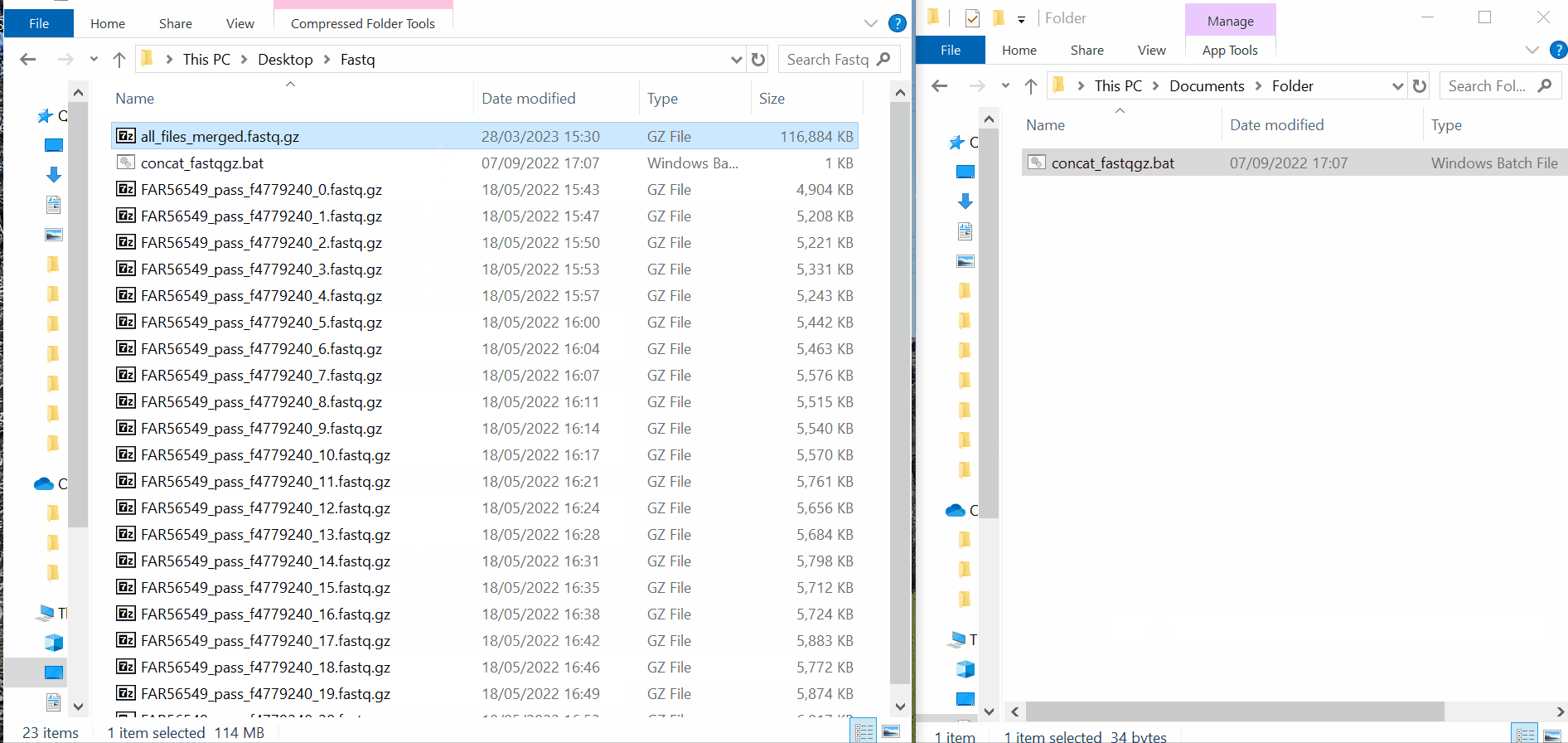
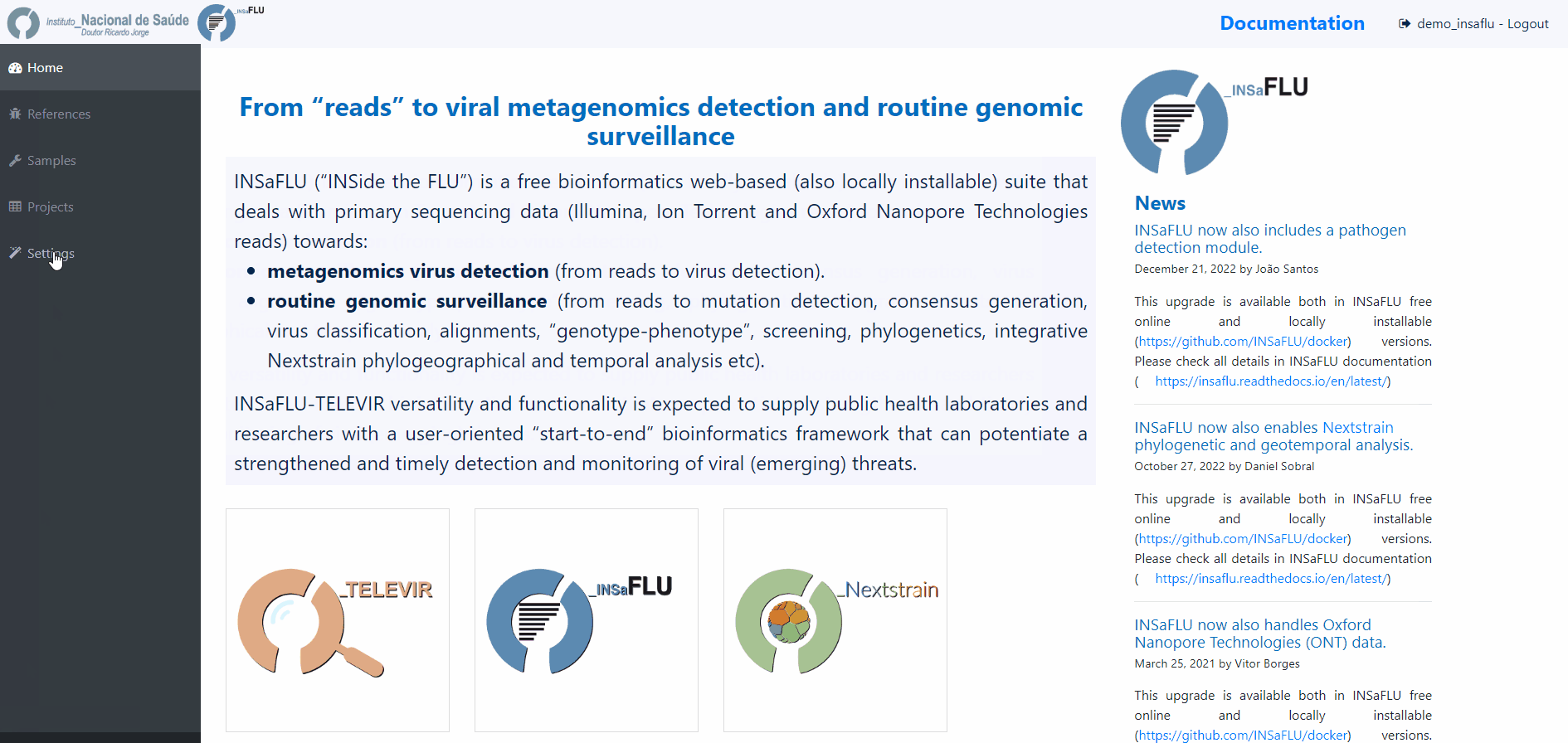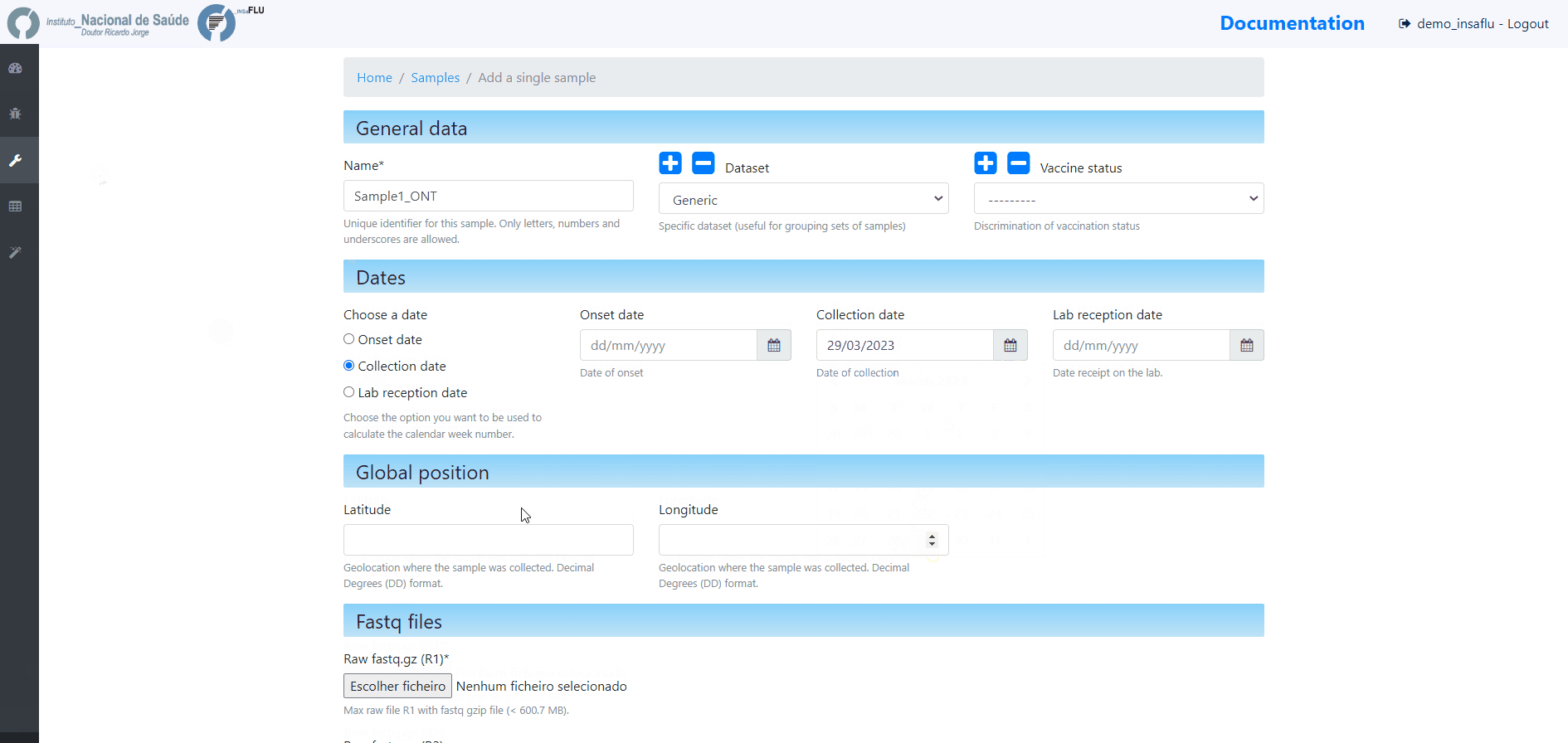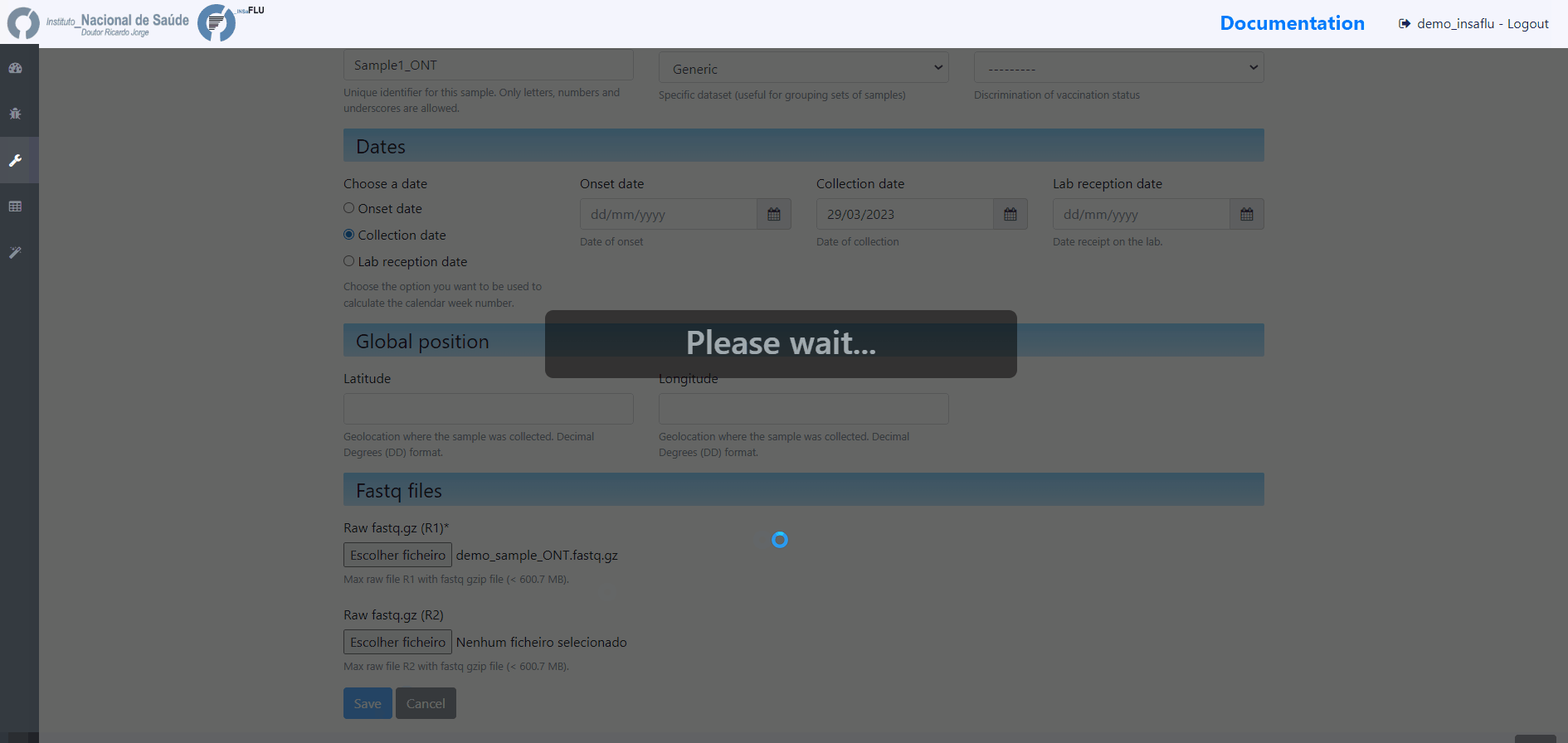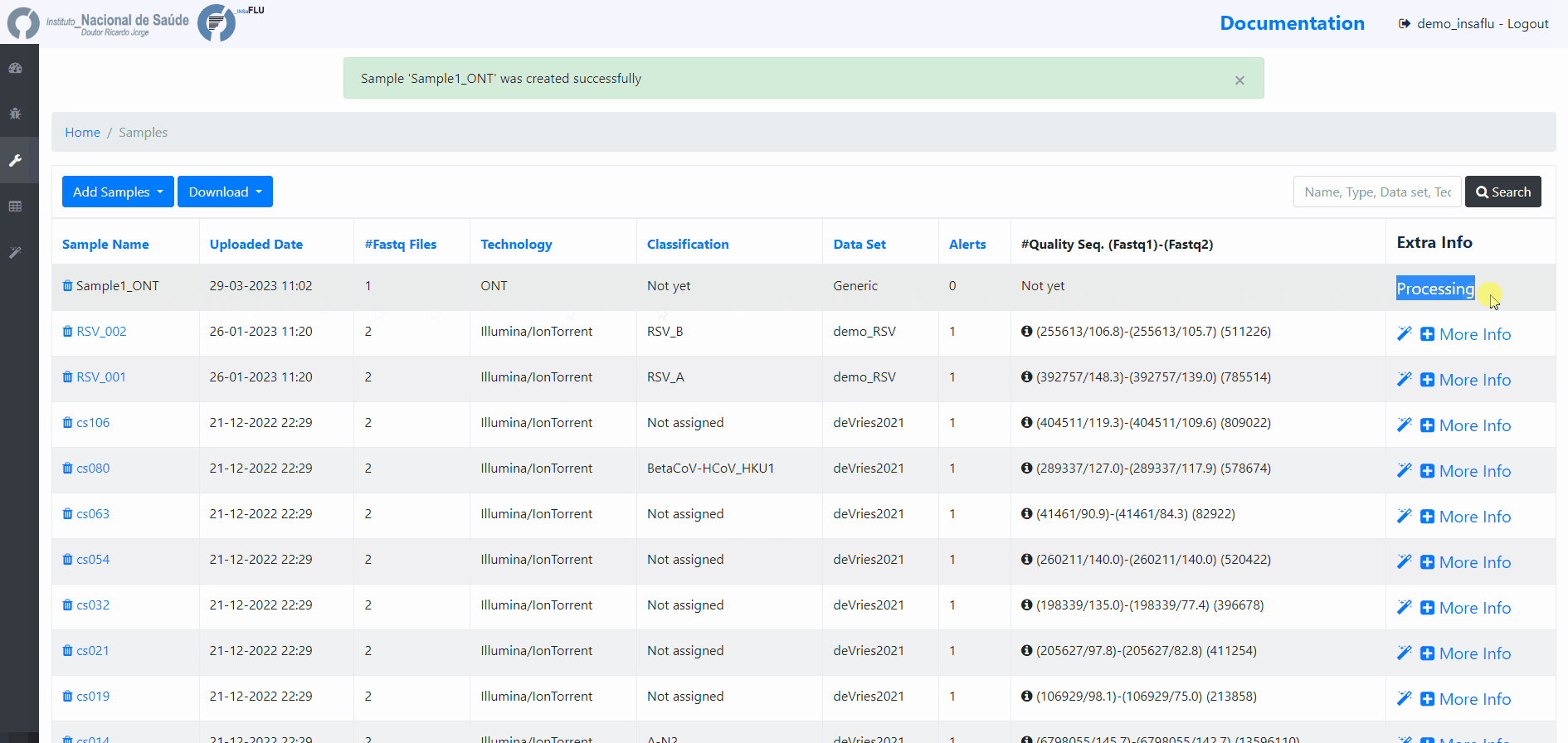
Samples’s metadata and respective NGS data (single-end or paired-end reads in fastq.gz format obtained through widely used technologies, such as Illumina or Ion Torrent) can be uploaded to INSaFLU as a batch (option 1) or individually (option 2):
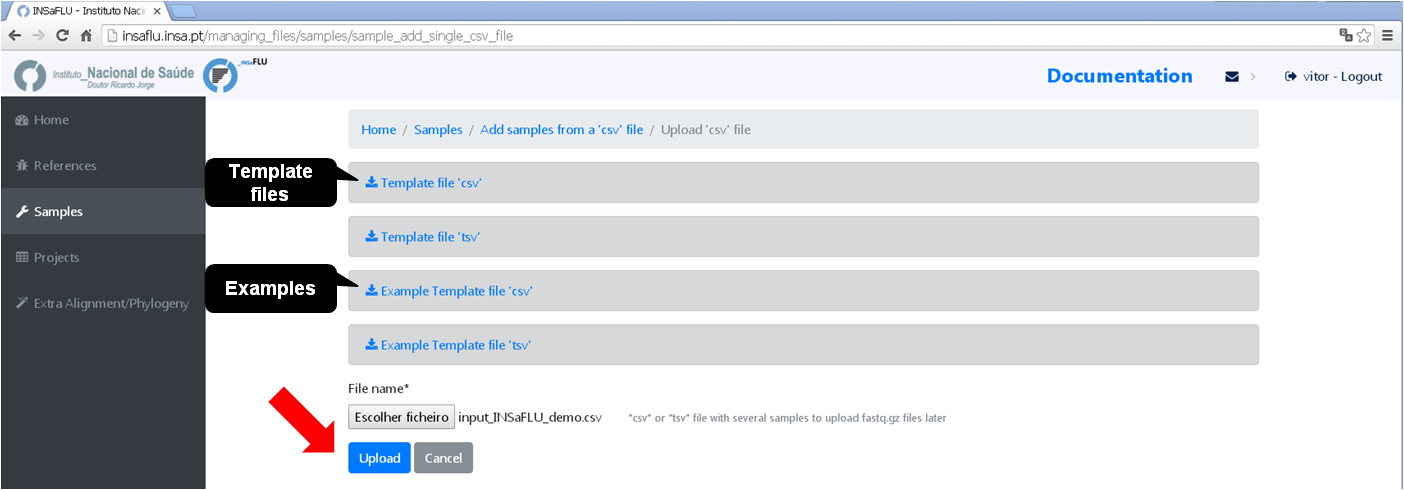
# Option 1 (Batch)
# Option 2 (Individual)
Updating Sample metadata
You can add/update the Samples’s metadata at any time by simply uploading a table comma-separated (.csv) or tab-separated (.tsv or .txt) table with the updated data (a template file is provided), as follows:
Go to Samples menu and choose Update metadata, then Load a new file, using one of the provided template files.
Important
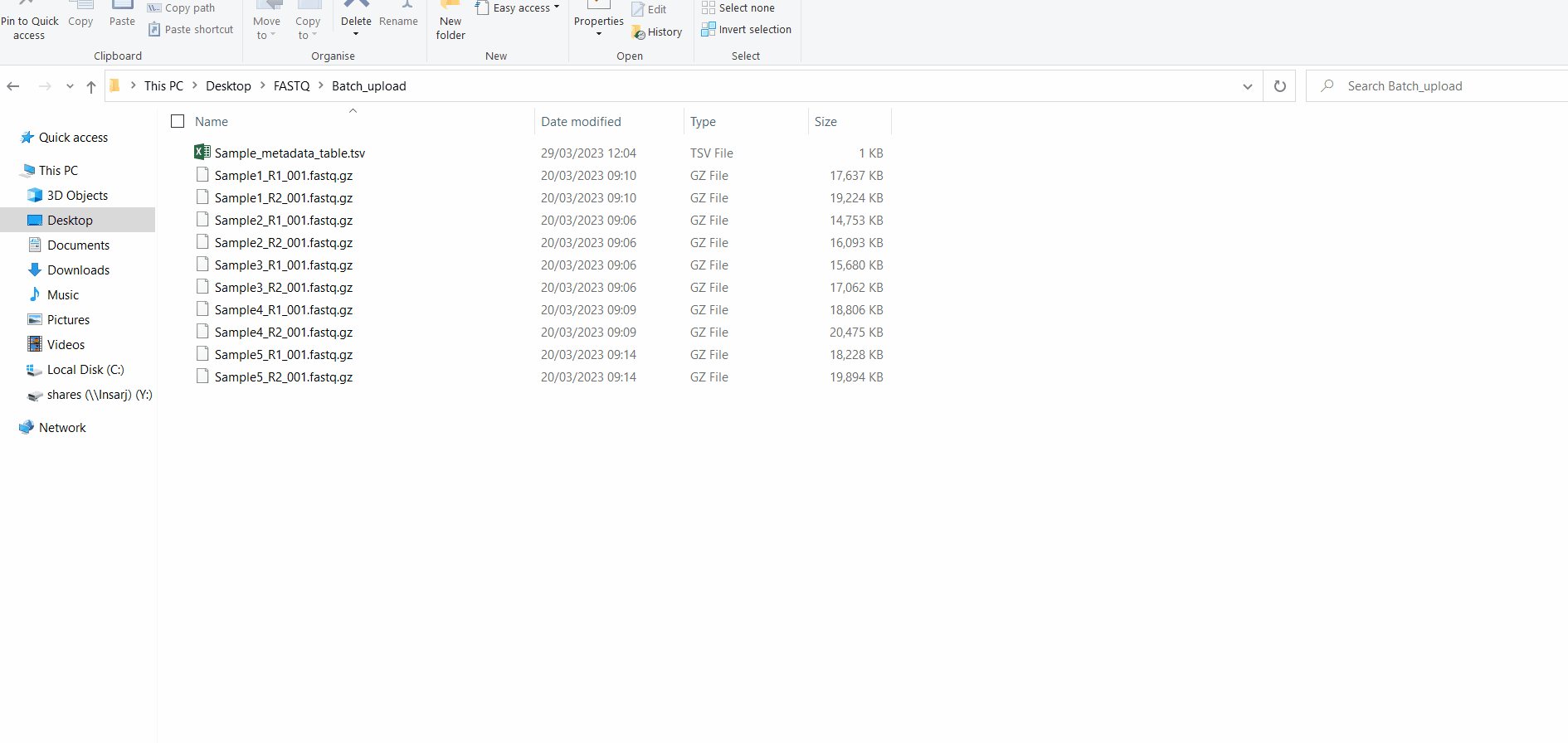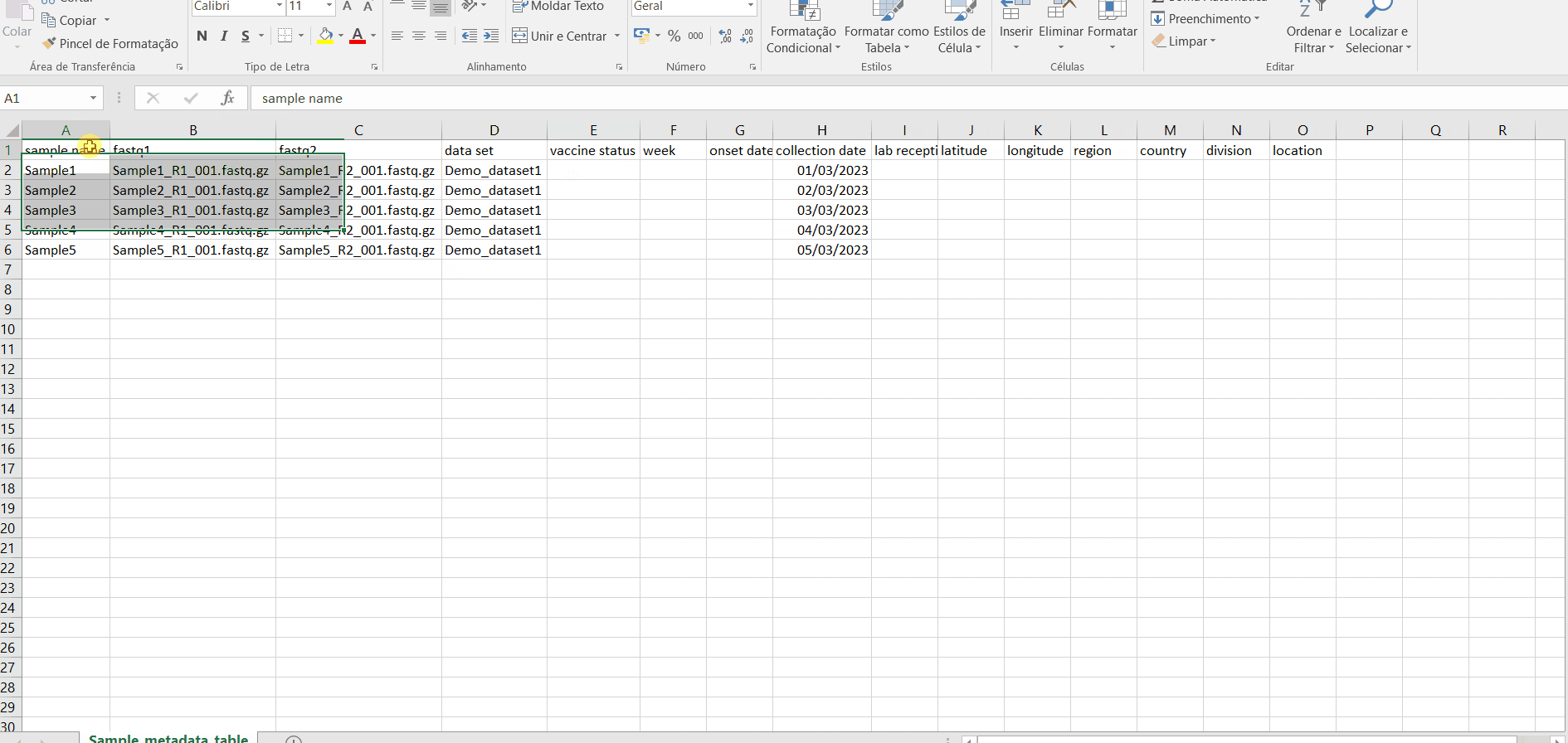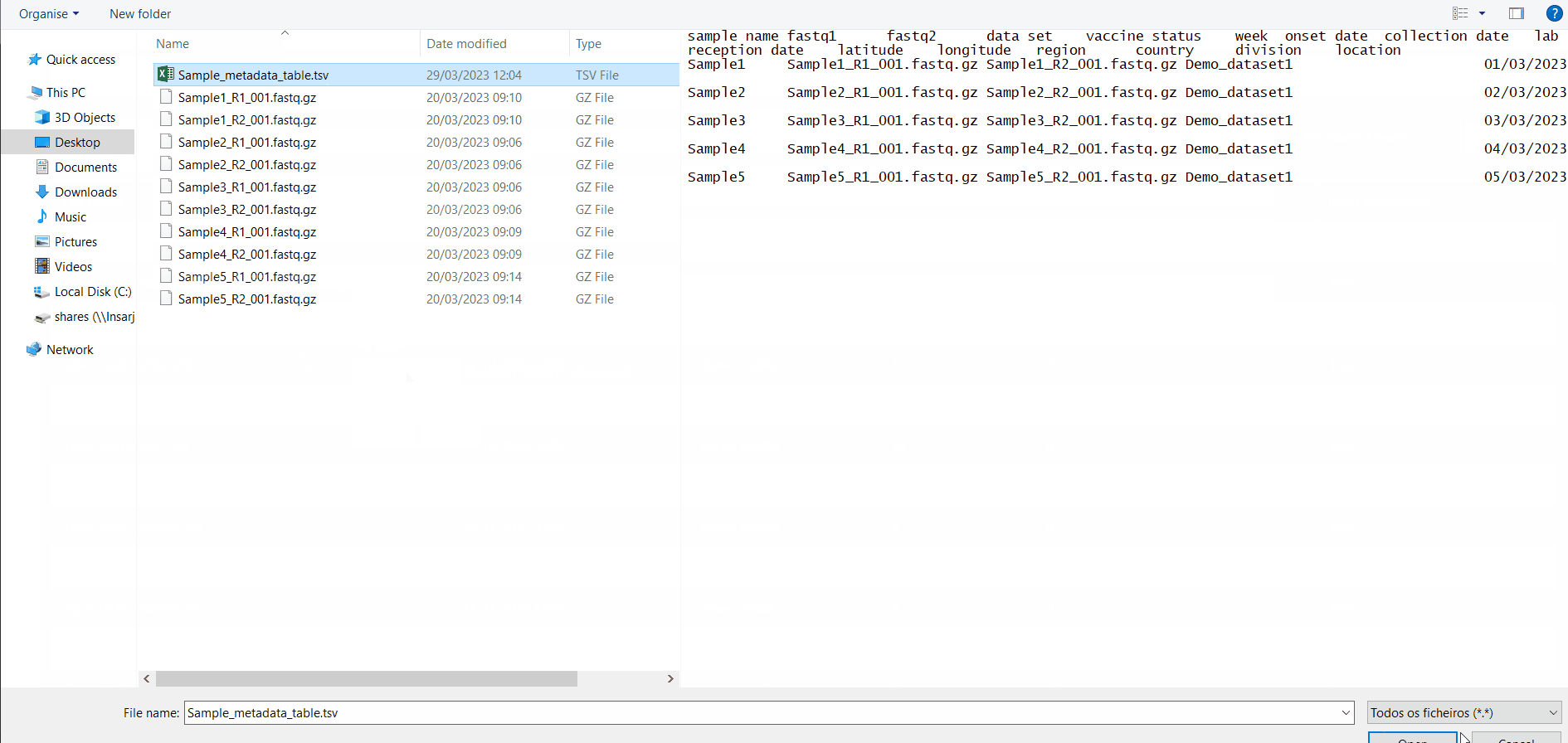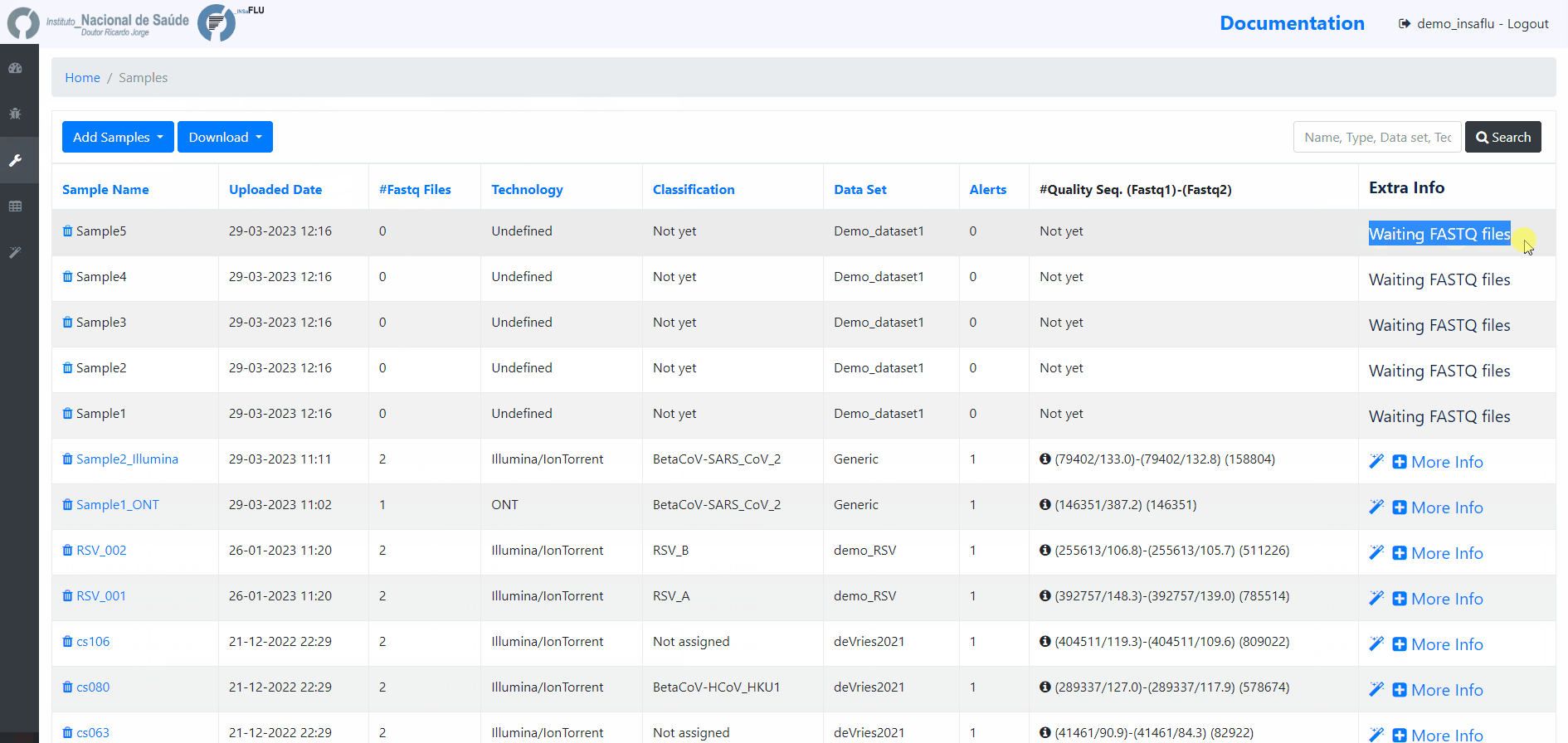
The table for Sample metadata update should contain the columns “sample name” (exactly corresponding to the samples to be updated), as well these additional variables (that may not be fulfilled): “data set”, ”vaccine status”, ”week”, ”onset date”, ”collection date”, ”lab reception date”, ”latitude”, ”longitude”, “country, region”, “division” and/or “location”. If you include data for “latitude” and “longitude” AND/OR country, region, division and/or location, these will be used to geographically locate your samples in the Nextstrain Datasets. Still, users are encouraged to include any other columns with metadata variables to be associated with samples (see advantages above).
Note
You cannot update the fastq files. To replace the fastq files associated with a given sample, you need to delete the respective sample and upload all data (metadata plus fastq) again, i.e., create a new sample. You must ensure that the sample to be deleted is not inserted in any Project before deleting it.
Samples names in your account must be unique and only numbers, letters and underscores are allowed.
You can add/update the Nextstrain metadata of a given Dataset by clicking in “Metadata for Nextstrain”, download the previous table, update it with new data and upload it again. Then, you need to click in the “hourglass” icon to Rebuild the Nextstrain outputs. When the new JSON file is generated, the updated metadata can then be drag-and drop to auspice.us after the JSON file to update the displayed metadata.
Notes: - This update option is exclusive of each Dataset. - To add/update the metadata associated with consensus sequences derived from read data in INSaFLU Projects module, you can also add/update the metadata following these instructions of the previous point “A” (this option is not available for “external” consensus sequences).
Uploading Reference data
INSaFLU needs reference sequence files to be used for reference-based mapping.
In References menu, INSaFLU provides a set of ready-to-use reference sequences, all publicly available at NCBI (or made available under permission of authors), currently including:
post-pandemic (2009) vaccine/reference influenza A(H1N1)pdm2009, A(H3N2) and B viruses (from both Northern and Southern hemispheres);
representative virus of multiple combinations of HA/NA subtypes (i.e., H1N1, H2N2, H5N1, H7N9, etc)
SARS-CoV-2 reference genome sequence (Wuhan-Hu-1; NCBI accession MN908947)
Mpox reference sequences
RSV references sequences
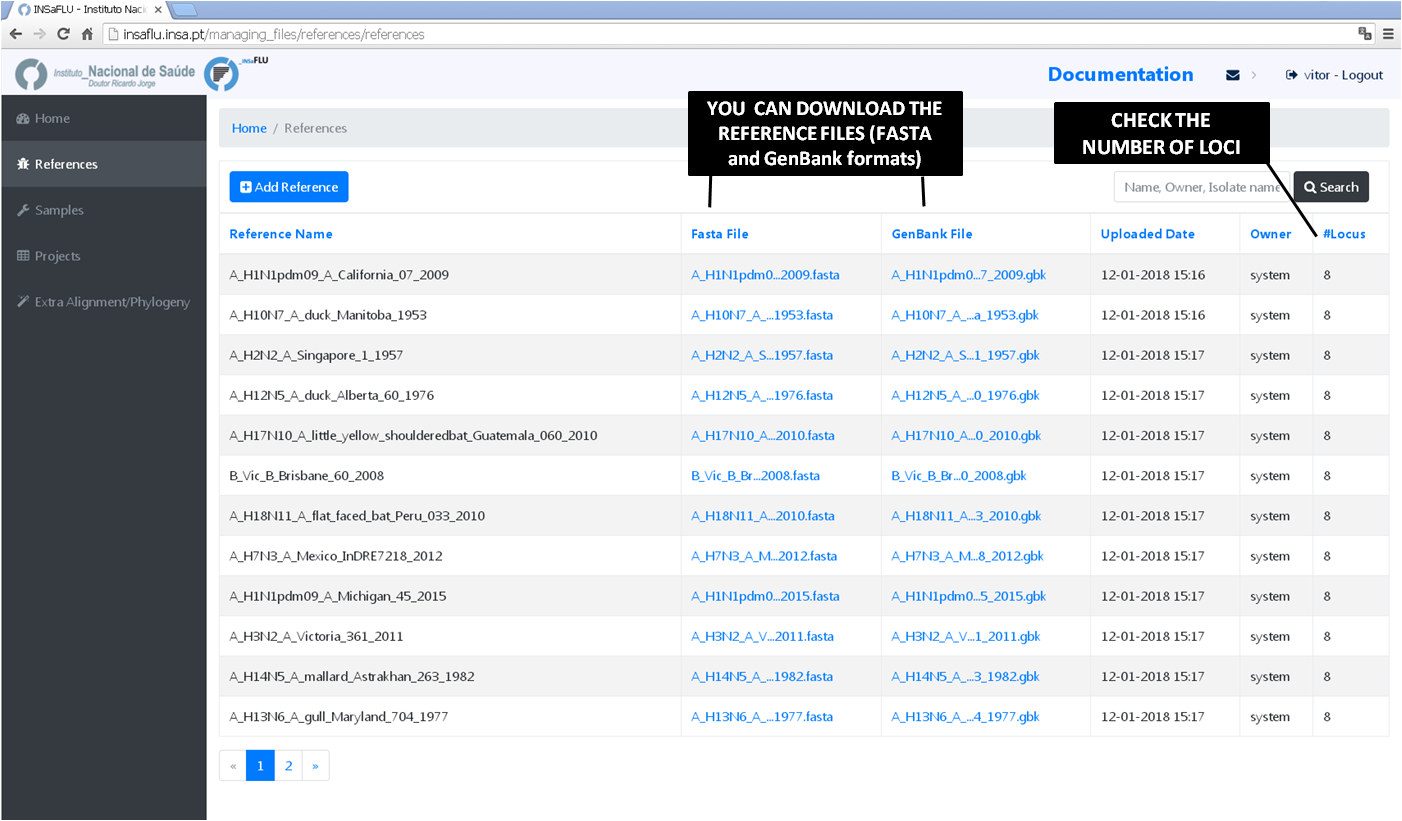
The current list of reference sequences, including loci size and NCBI accession numbers is provided here:
INSaFLU_current_REFERENCE_DATABASE_07_03_2023.xlsx
NOTE: The default seasonal influenza reference files (FASTA and GenBank formats) have been prepared to fit amplicon-based schemas capturing the whole CDS of the main eight genes of influenza virus (PB2, PB1, PA, HA, NP, NA, M and NS), such as the wet-lab pre-NGS protocol for influenza whole genome amplification adapted from a RT-PCR assay described by Zhou and colleagues (Zhou et al, 2009, for Influenza A; and Zhou et al, 2014, for Influenza B; Zhou and Wentworth, 2012).
You can download the suggested protocol here: Suggested_RT_PCR_assay_for_influenza_WGS.pdf
Important
NO FURTHER ACTIONS ARE NEEDED if you are using the suggested wet-lab pre-NGS protocol and you want to compare your sequences against a reference available at INSaFLU database.
However, you may need to UPLOAD additional reference files to the user-restricted reference database. For instance, you may need to upload the A/H3N2 vaccine reference sequence for the season 2017/2018 (A/Hong Kong/4801/2014 virus), which is not freely available.
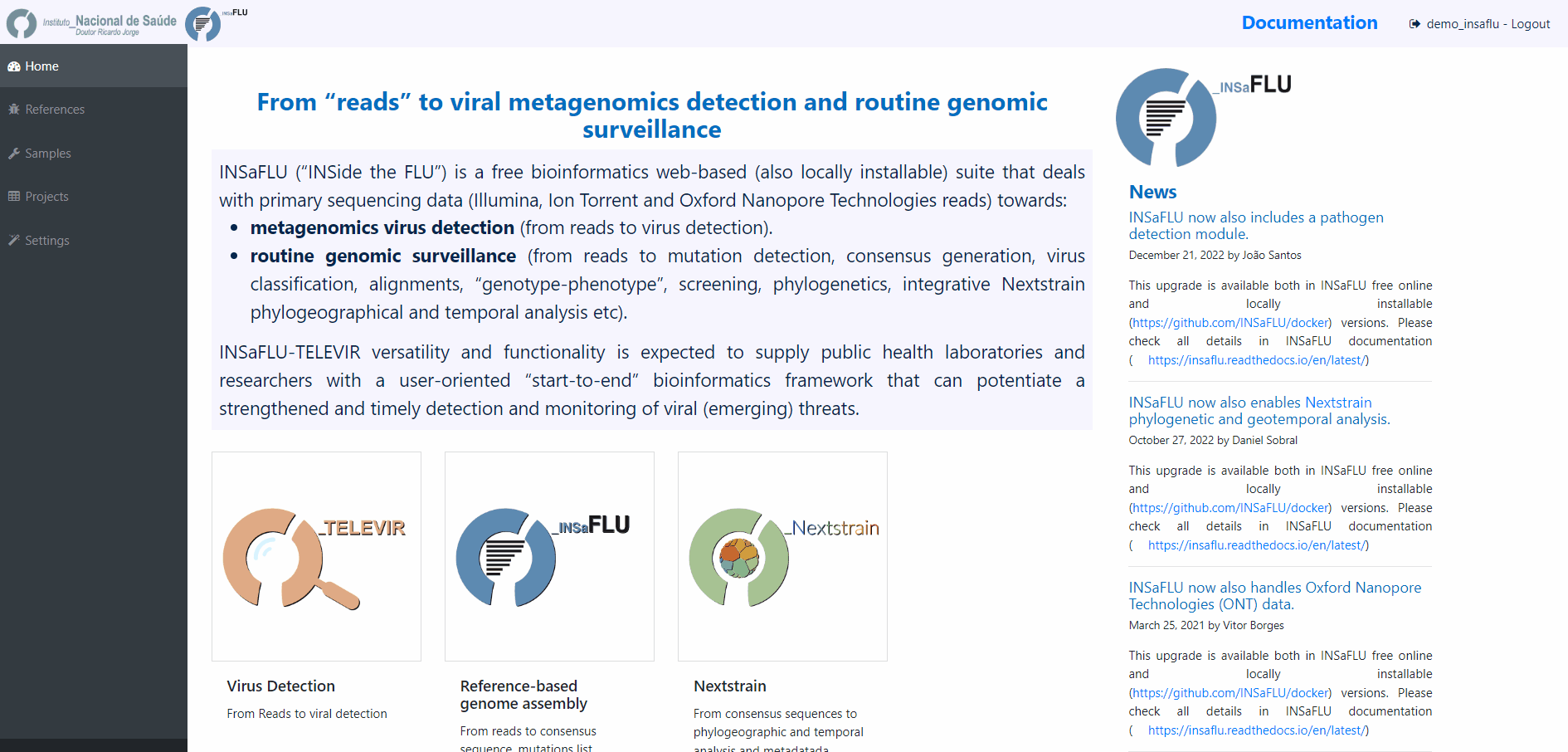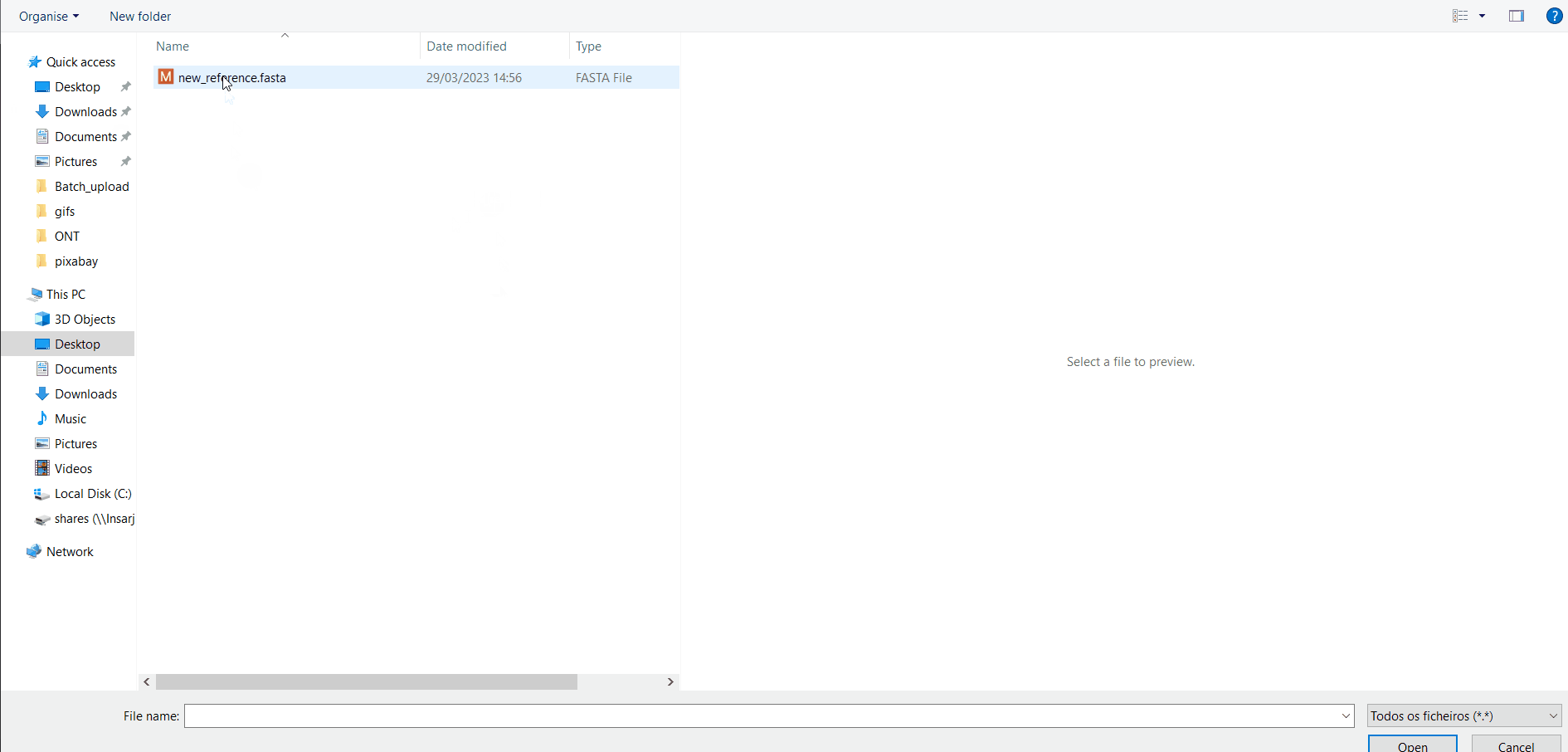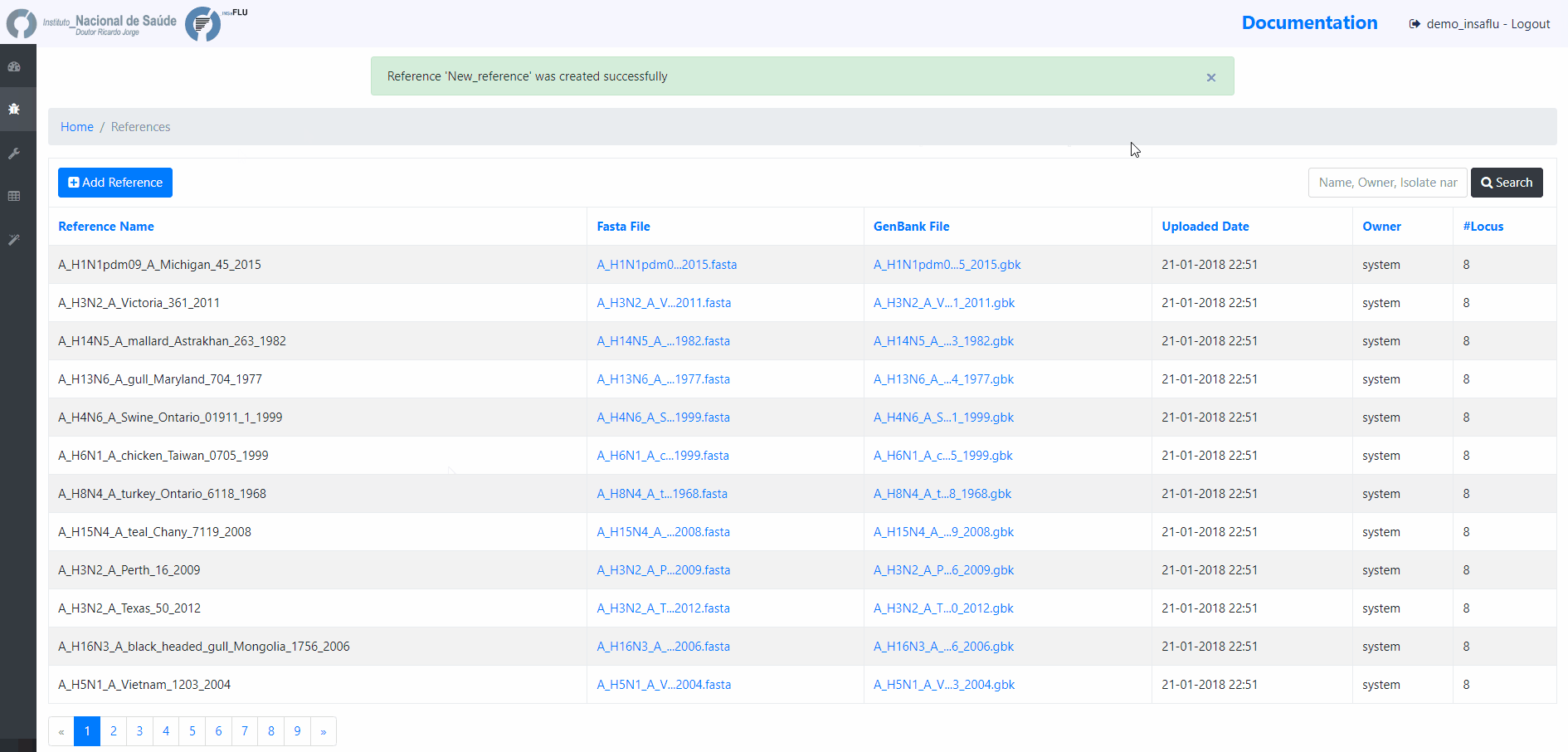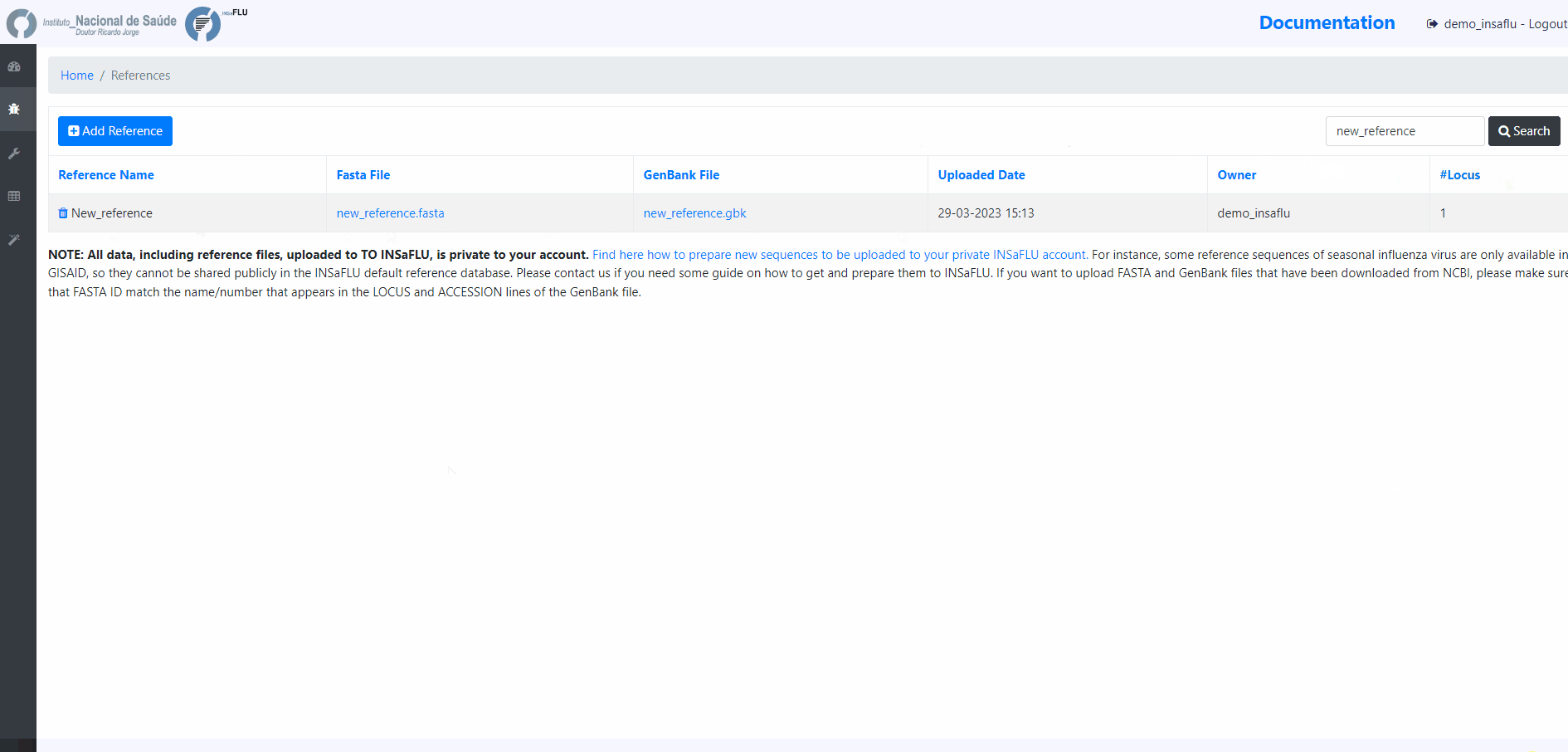
To upload additional reference sequences, GO TO References MENU and CHOOSE Add Reference
You can eitheir:
Upload a FASTA + GENBANK file

Note
Your genbank file must be compatible with INSaFLU. ## See below a GUIDE to generate additional reference sequences
Upload a FASTA file
In this case, INSaFLU will automatically generate an annotation in GenBabk format*
Note
Important notes - upload new References:
Multi-FASTA files to be uploaded will typically contain the set of reference sequences that constitute the influenza “whole-genome” sequence of a particular virus (e.g, the combination of the traditional 8 amplicons targeting the 8 eight influenza RNA segments). Still, you are free to upload references files including a specific panel of segments/genes (e.g, segments 4 and 6, which encode the surface proteins HA and NA, respectively)
Each individual sequence of the multi-FASTA file should ideally have the size of each “intra-amplicon” target sequence that you capture by each one of the RT-PCR amplicons. Otherwise, you will get regions with no or low coverage (these will be masked with undefined bases NNN according to the user-defined coverage thresholds).
INSaFLU automatically annotates the uploaded multi-FASTA sequences upon submission, but, if you prefer, you can also upload (optionally) the respective multi-GenBank file.
## See below a GUIDE to generate additional reference sequences
GUIDE TO GENERATE ADDITIONAL REFERENCE SEQUENCES
Please take this guide into account when generating additional reference sequences.
(Multi-)FASTA files to be upload typically contain the reference sequence(s) that constitute the “whole-genome” sequence of a particular virus (e.g, the combination of the traditional 8 amplicons targeting the 8 eight influenza RNA segments in a MULTI-FASTA file or the reference genome sequence for SARS-CoV2 in a Single FASTA file). Each individual sequence should ideally have the precise size of each “intra-amplicon” target sequence that you capture by each one of the RT-PCR amplicons. Otherwise, you will obtain regions with no or low coverage (these will be masked with undefined bases NNN according to the user-defined coverage thresholds)
You may generate your (multi-)FASTA files in order to fit your amplicon schema by simply adjusting the whole-genome sequences available for download at INSaFLU or at influenza-specific sequence repositories, such as the Influenza Research Database (https://www.fludb.org), NCBI Influenza Virus Resource (https://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi?go=database) and EpiFLU/GISAID (https://www.gisaid.org/).
An easy way to handle/generate (multi-)FASTA files is by opening a text file (e.g., NOTEPAD) and paste individual sequences after each header line. The FASTA IDs (after the ‘>’ character) represent the individual sequence names. For the sake of simplicity, you may designate each sequence as 1, 2, 3, 4, 5, 6 , 7 and 8 (see example), following the traditional influenza segments order (keeping this numerical order is advisable). At the end, you just have to save the (multi-)FASTA file as “.fasta”. Please avoid symbols or blank spaces in the file names.
example:
A_H3N2_A_Perth_16_2009.fastaINSaFLU automatically annotates uploaded (multi-)FASTA sequences upon submission, but, if you prefer, you can also upload (optionally) the respective multi-GenBank file. If you upload FASTA and respective GenBank files that have been downloaded from NCBI, please make sure that FASTA ID(s) (after the ‘>’ character) match the name/number that appears in the LOCUS and ACCESSION lines of the GenBank file..
INSaFLU requires reference sequences exclusively composed by non-degenerate bases (i.e. A, T, C, or G). As such, please ensure that all degenerated bases (e.g., R, Y, M, K, S and W) are replaced by non-degenerate sequences before uploading. The choice of the base used in the replacement (e.g., “A” or “G” when replacing an “R”) has no impact on the analysis. It simply means that mutations falling in the replaced nucleotide position will be reported taking into account the reference base selected.
IMPORTANT:
The influenza GenBank files available at NCBI are not compatible with the “HA1 numbering” (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4229193/) commonly applied for seasonal influenza. As such, in order to get mutations annotated for each peptide (signal peptide, HA1 and HA2 peptides), instead of the full-protein, make sure you modified the GenBank file as follows:
THESE ORIGINAL LINES in THE GENBANK FILE ….
…SHOULD BE REPLACED BY:
Explore your Sample and Reference databases
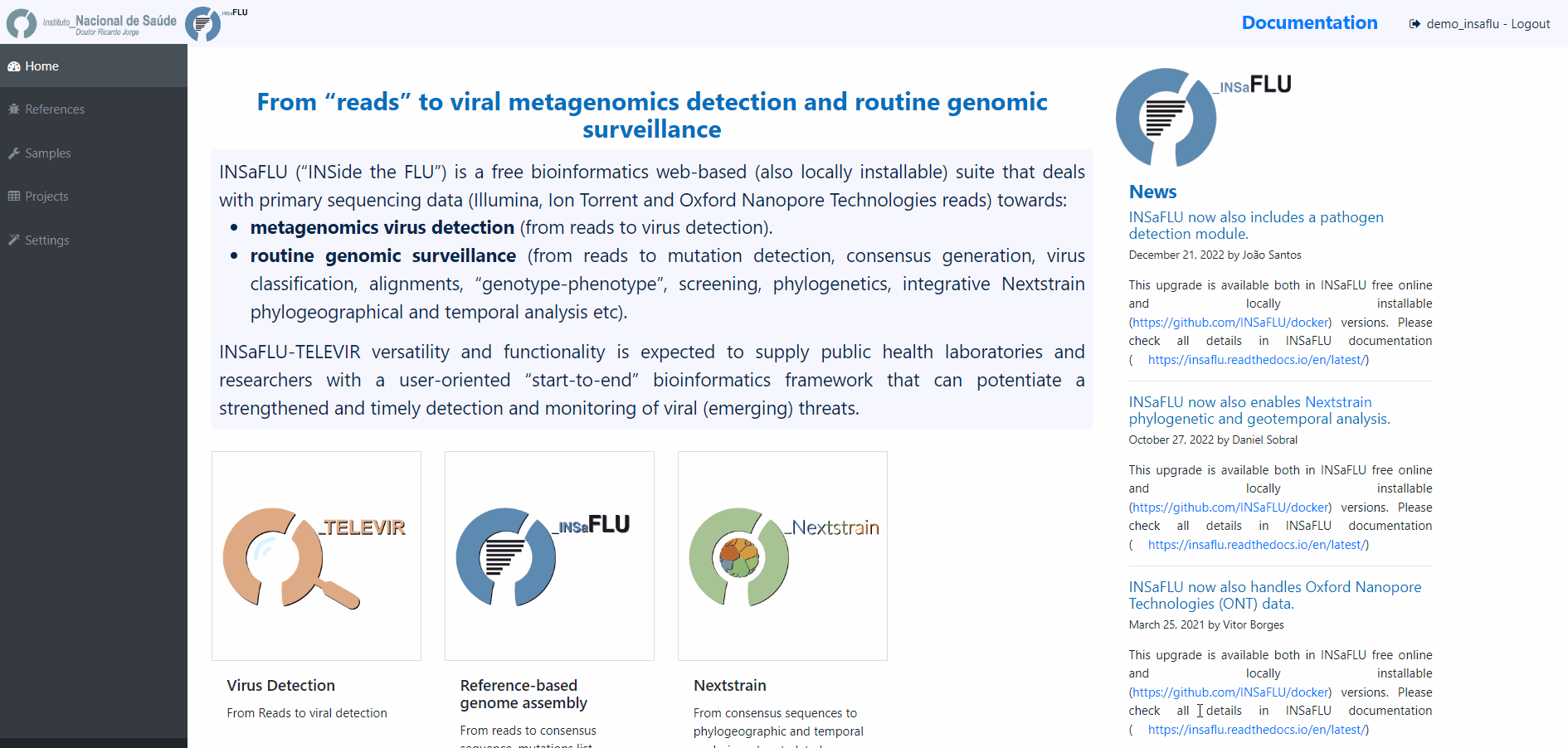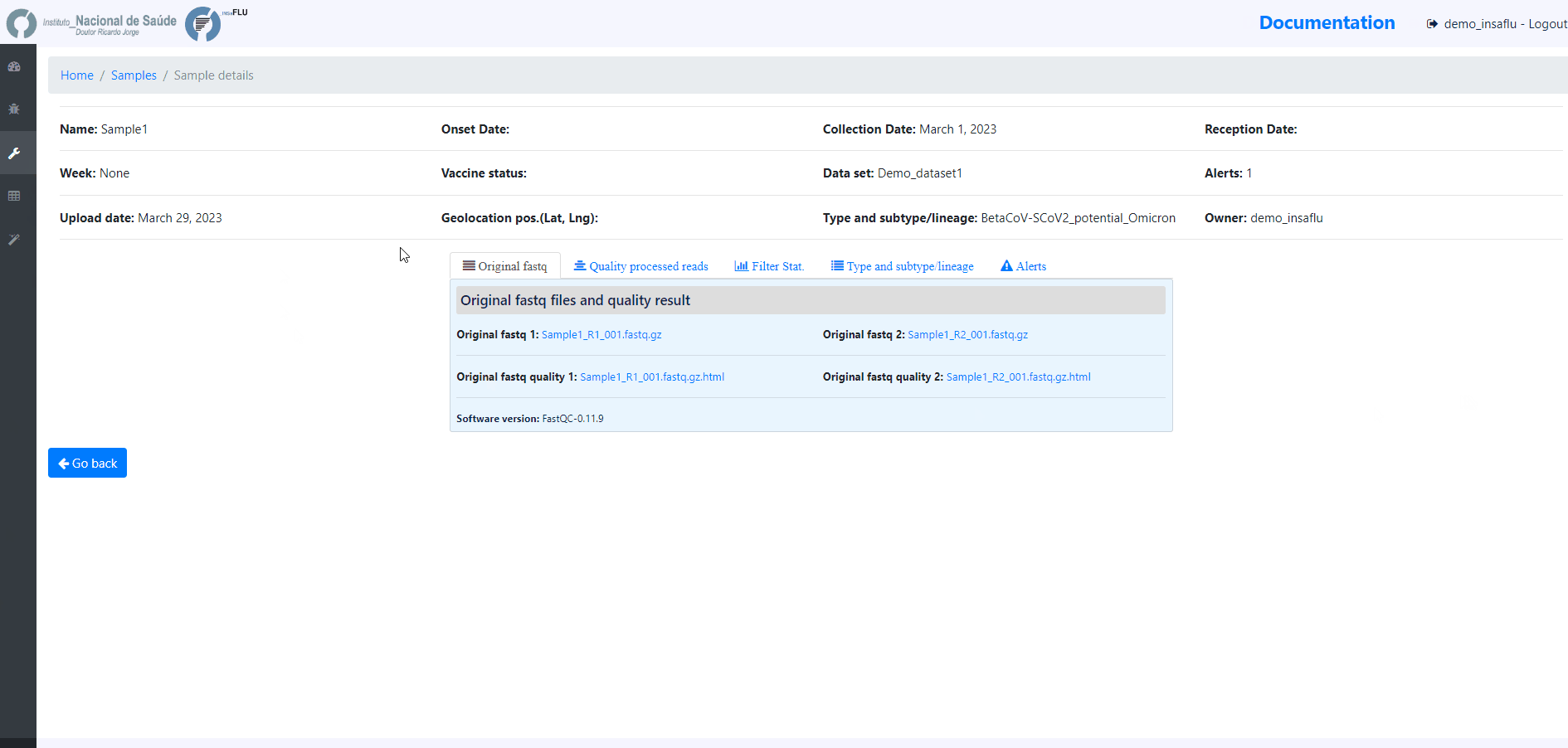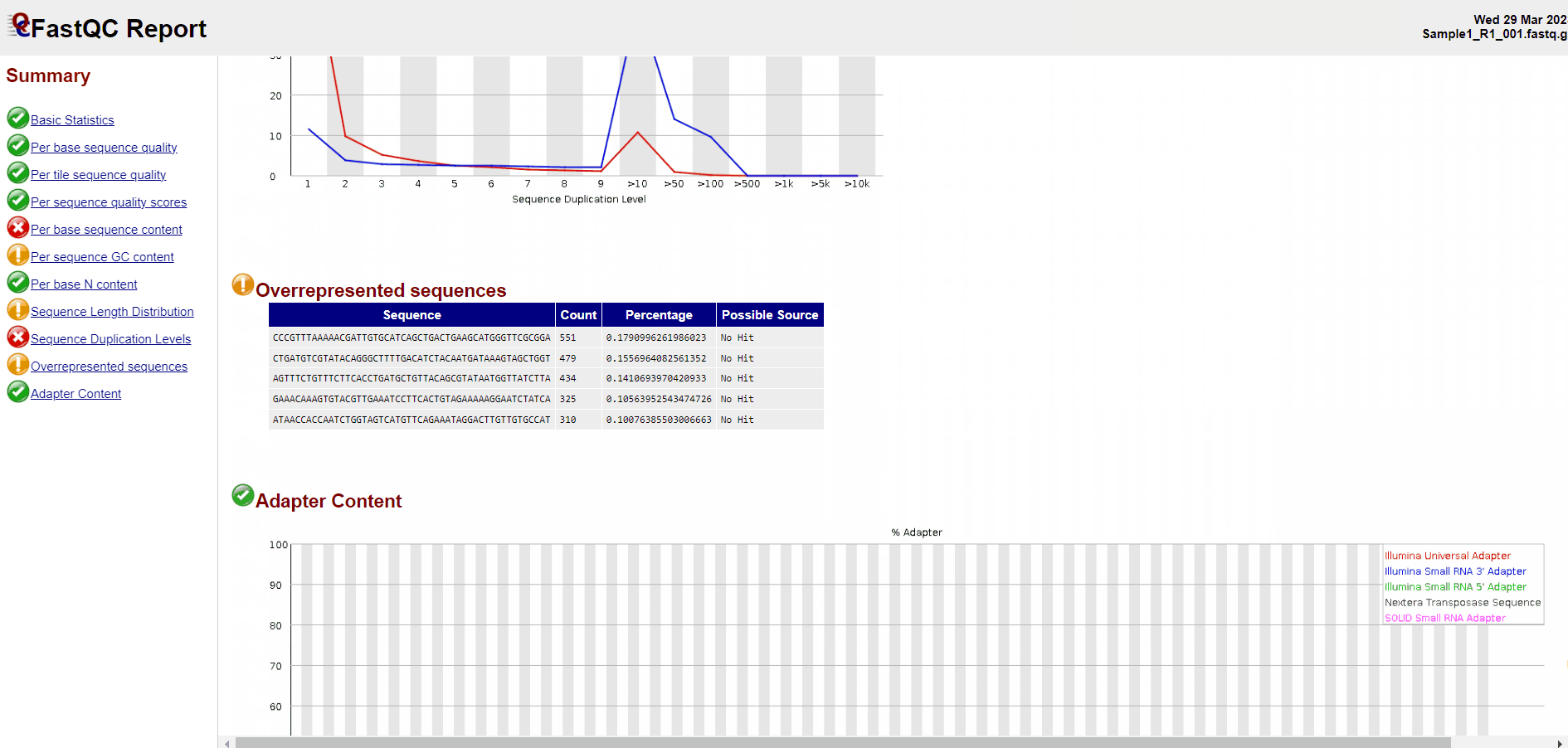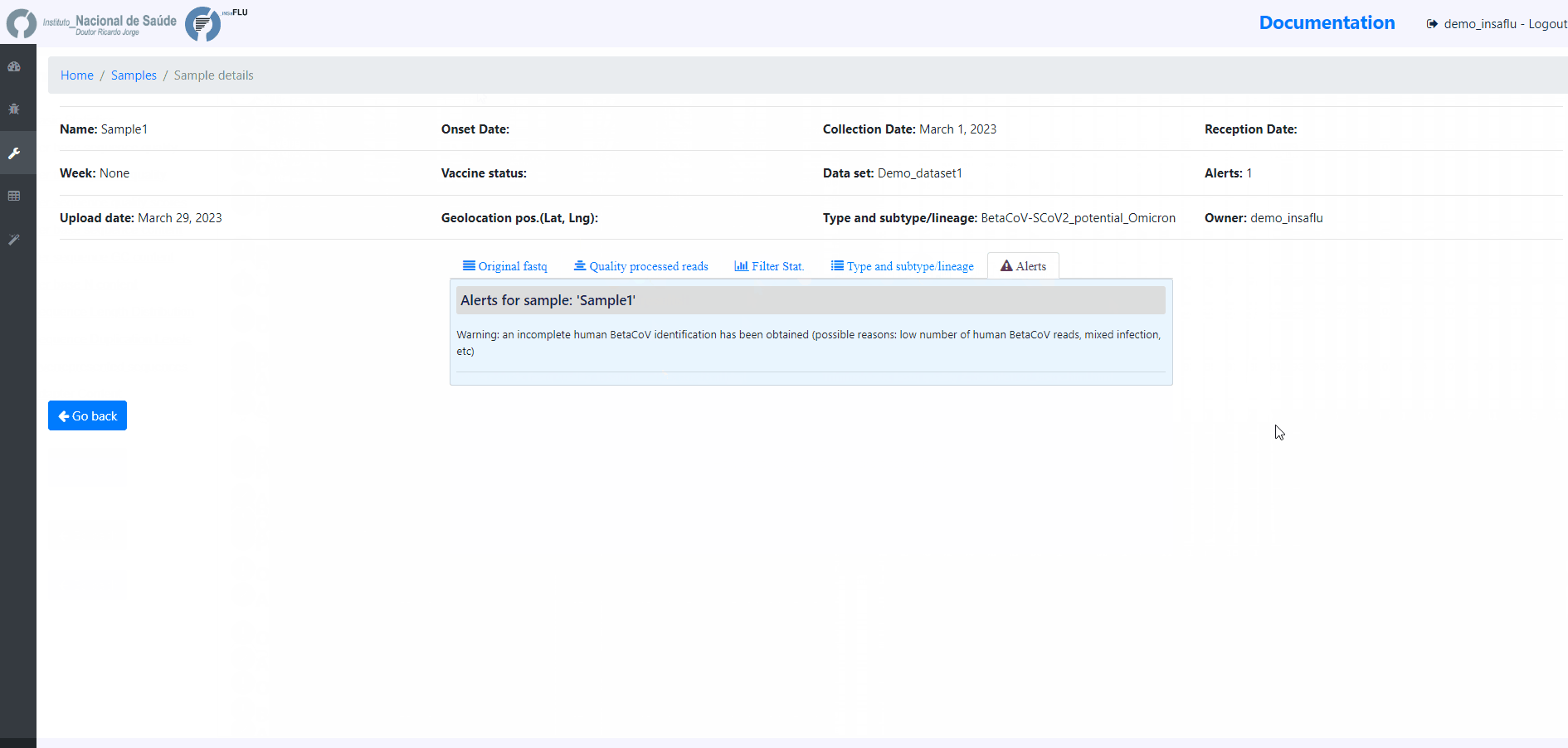
Samples menu displays all information for all loaded samples (Samples’ names in your account must be unique).
Upon submission, INSaFLU automatically updates samples’ information with reads quality and typing data (automate bioinformatics pipeline modules “Read quality analysis and improvement” and Type and sub-type detection”; see Data analysis in the Documentation).
Just explore the “More info” icon next to each sample.
References menu displays all information for all reference sequences available at your confidential session.
Both FASTA and GenBank files can be downloaded by clicking on the displayed links.

References:
Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, Wentworth DE. 2009. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J Virol, 83:10309-13.
Zhou B, Lin X, Wang W, Halpin RA, Bera J, Stockwell TB, Barr IG, Wentworth DE. 2014. Universal influenza B virus genomic amplification facilitates sequencing, diagnostics, and reverse genetics. J Clin Microbiol, 52:1330-1337.
Zhou B, Wentworth DE. 2012. Influenza A virus molecular virology techniques. Methods Mol Biol, 865:175-92.